The LG/J murine strain exhibits near-normal tendon biomechanical properties following a full-length central patellar tendon defect
- PMID: 27552106
- PMCID: PMC10552235
- DOI: 10.1080/03008207.2016.1213247
The LG/J murine strain exhibits near-normal tendon biomechanical properties following a full-length central patellar tendon defect
Abstract
Purpose of the study: Identifying biological success criteria is needed to improve therapies, and one strategy for identifying them is to analyze the RNA transcriptome for successful and unsuccessful models of tendon healing. We have characterized the MRL/MpJ murine strain and found improved mechanical outcomes following a central patellar tendon (PT) injury. In this study, we evaluate the healing of the LG/J murine strain, which comprises 75% of the MRL/MpJ background, to determine if the LG/J also exhibits improved biomechanical properties following injury and to determine differentially expressed transcription factors across the C57BL/6, MRL/MpJ and the LG/J strains during the early stages of healing.
Materials and methods: A full-length, central PT defect was created in 16-20 week old MRL/MpJ, LG/J, and C57BL/6 murine strains. Mechanical properties were assessed at 2, 5, and 8 weeks post surgery. Transcriptomic expression was assessed at 3, 7, and 14 days following injury using a novel clustering software program to evaluate differential expression of transcription factors.
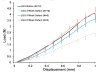
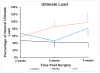
Results: Average LG/J structural properties improved to 96.7% and 97.2% of native LG/J PT stiffness and ultimate load by 8 weeks post surgery, respectively. We found the LG/J responded by increasing expression of transcription factors implicated in the inflammatory response and collagen fibril organization.
Conclusions: The LG/J strain returns to normal structural properties by 8 weeks, with steadily increasing properties at each time point. Future work will characterize the cell populations responding to injury and investigate the role of the differentially expressed transcription factors during healing.
Keywords: Biomechanical properties; mouse model; tendon healing; transcription factors; transcriptome.
Conflict of interest statement
The authors declare they have no actual or competing financial interests.
Figures










References
-
- The burden of musculoskeletal diseases in the united states (BMUS) 3. American Academy of Orthopaedic Surgeons;; 2014.
-
- Awad HA, Boivin GP, Dressler MR, Smith FNL, Young RG, Butler DL. Repair of patellar tendon injuries using a cell-collagen composite. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2003;21:420–31. - PubMed
-
- Dyment NA, Kazemi N, Aschbacher-Smith LE, Barthelery NJ, Kenter K, Gooch C, Shearn JT, Wylie C, Butler DL. The relationships among spatiotemporal collagen gene expression, histology, and biomechanics following full-length injury in the murine patellar tendon. Journal of Orthopaedic Research. 2012;30:28–36. - PMC - PubMed
-
- Bruns J, Kampen J, Kahrs J, Plitz W. Achilles tendon rupture: Experimental results on spontaneous repair in a sheep-model. Knee Surgery Sports Traumatology Arthroscopy. 2000;8:364–369. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
