Small Molecule Inhibitor of NRF2 Selectively Intervenes Therapeutic Resistance in KEAP1-Deficient NSCLC Tumors
- PMID: 27552339
- PMCID: PMC5367156
- DOI: 10.1021/acschembio.6b00651
Small Molecule Inhibitor of NRF2 Selectively Intervenes Therapeutic Resistance in KEAP1-Deficient NSCLC Tumors
Abstract
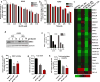
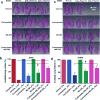
Loss of function mutations in Kelch-like ECH Associated Protein 1 (KEAP1), or gain-of-function mutations in nuclear factor erythroid 2-related factor 2 (NRF2), are common in non-small cell lung cancer (NSCLC) and associated with therapeutic resistance. To discover novel NRF2 inhibitors for targeted therapy, we conducted a quantitative high-throughput screen using a diverse set of ∼400 000 small molecules (Molecular Libraries Small Molecule Repository Library, MLSMR) at the National Center for Advancing Translational Sciences. We identified ML385 as a probe molecule that binds to NRF2 and inhibits its downstream target gene expression. Specifically, ML385 binds to Neh1, the Cap 'N' Collar Basic Leucine Zipper (CNC-bZIP) domain of NRF2, and interferes with the binding of the V-Maf Avian Musculoaponeurotic Fibrosarcoma Oncogene Homologue G (MAFG)-NRF2 protein complex to regulatory DNA binding sequences. In clonogenic assays, when used in combination with platinum-based drugs, doxorubicin or taxol, ML385 substantially enhances cytotoxicity in NSCLC cells, as compared to single agents. ML385 shows specificity and selectivity for NSCLC cells with KEAP1 mutation, leading to gain of NRF2 function. In preclinical models of NSCLC with gain of NRF2 function, ML385 in combination with carboplatin showed significant antitumor activity. We demonstrate the discovery and validation of ML385 as a novel and specific NRF2 inhibitor and conclude that targeting NRF2 may represent a promising strategy for the treatment of advanced NSCLC.
Figures





References
-
- American Cancer Society. Cancer Facts and Figures 2013. Atlanta, Ga: American Cancer Society; 2013.
-
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. - PubMed
-
- Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW, Wakabayashi N, Dewi R, Boros LG, Gonzalez FJ, Gabrielson E, Wong KK, Girnun G, Biswal S. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest. 2013;123:2921–2934. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

