Diagnostic Test Accuracy of a 2-Transcript Host RNA Signature for Discriminating Bacterial vs Viral Infection in Febrile Children
- PMID: 27552617
- PMCID: PMC5997174
- DOI: 10.1001/jama.2016.11236
Diagnostic Test Accuracy of a 2-Transcript Host RNA Signature for Discriminating Bacterial vs Viral Infection in Febrile Children
Erratum in
-
Errors in Text, Figure, and Online Supplement.JAMA. 2017 Feb 7;317(5):538. doi: 10.1001/jama.2016.18601. JAMA. 2017. PMID: 28170459 No abstract available.
Abstract
Importance: Because clinical features do not reliably distinguish bacterial from viral infection, many children worldwide receive unnecessary antibiotic treatment, while bacterial infection is missed in others.
Objective: To identify a blood RNA expression signature that distinguishes bacterial from viral infection in febrile children.
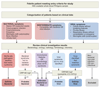
Design, setting, and participants: Febrile children presenting to participating hospitals in the United Kingdom, Spain, the Netherlands, and the United States between 2009-2013 were prospectively recruited, comprising a discovery group and validation group. Each group was classified after microbiological investigation as having definite bacterial infection, definite viral infection, or indeterminate infection. RNA expression signatures distinguishing definite bacterial from viral infection were identified in the discovery group and diagnostic performance assessed in the validation group. Additional validation was undertaken in separate studies of children with meningococcal disease (n = 24) and inflammatory diseases (n = 48) and on published gene expression datasets.
Exposures: A 2-transcript RNA expression signature distinguishing bacterial infection from viral infection was evaluated against clinical and microbiological diagnosis.
Main outcomes and measures: Definite bacterial and viral infection was confirmed by culture or molecular detection of the pathogens. Performance of the RNA signature was evaluated in the definite bacterial and viral group and in the indeterminate infection group.
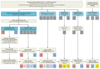
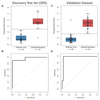
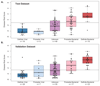
Results: The discovery group of 240 children (median age, 19 months; 62% male) included 52 with definite bacterial infection, of whom 36 (69%) required intensive care, and 92 with definite viral infection, of whom 32 (35%) required intensive care. Ninety-six children had indeterminate infection. Analysis of RNA expression data identified a 38-transcript signature distinguishing bacterial from viral infection. A smaller (2-transcript) signature (FAM89A and IFI44L) was identified by removing highly correlated transcripts. When this 2-transcript signature was implemented as a disease risk score in the validation group (130 children, with 23 definite bacterial, 28 definite viral, and 79 indeterminate infections; median age, 17 months; 57% male), all 23 patients with microbiologically confirmed definite bacterial infection were classified as bacterial (sensitivity, 100% [95% CI, 100%-100%]) and 27 of 28 patients with definite viral infection were classified as viral (specificity, 96.4% [95% CI, 89.3%-100%]). When applied to additional validation datasets from patients with meningococcal and inflammatory diseases, bacterial infection was identified with a sensitivity of 91.7% (95% CI, 79.2%-100%) and 90.0% (95% CI, 70.0%-100%), respectively, and with specificity of 96.0% (95% CI, 88.0%-100%) and 95.8% (95% CI, 89.6%-100%). Of the children in the indeterminate groups, 46.3% (63/136) were classified as having bacterial infection, although 94.9% (129/136) received antibiotic treatment.
Conclusions and relevance: This study provides preliminary data regarding test accuracy of a 2-transcript host RNA signature discriminating bacterial from viral infection in febrile children. Further studies are needed in diverse groups of patients to assess accuracy and clinical utility of this test in different clinical settings.
Conflict of interest statement
Figures

Comment in
-
Genetics and the Evaluation of the Febrile Child.JAMA. 2016 Aug 23-30;316(8):824-5. doi: 10.1001/jama.2016.11137. JAMA. 2016. PMID: 27552615 No abstract available.
-
RNA signature test to distinguish bacterial from viral infection.J Pediatr. 2017 Mar;182:401-404. doi: 10.1016/j.jpeds.2016.12.066. J Pediatr. 2017. PMID: 28237452 No abstract available.
-
Diagnosis of Bacterial Infection Using a 2-Transcript Host RNA Signature in Febrile Infants 60 Days or Younger.JAMA. 2017 Apr 18;317(15):1577-1578. doi: 10.1001/jama.2017.1365. JAMA. 2017. PMID: 28418473 No abstract available.
References
-
- Iroh Tam PY, Bernstein E, Ma X, Ferrieri P. Blood culture in evaluation of pediatric community-acquired Pneumonia: a systematic review and meta-analysis. Hosp Pediatr. 2015;5(6):324–336. - PubMed
-
- Martin NG, Sadarangani M, Pollard AJ, Goldacre MJ. Hospital admission rates for meningitis and septicaemia caused by Haemophilus influenzae, Neisseria meningitidis, and Streptococcus pneumoniae in children in England over five decades: a population-based observational study. The Lancet Infectious diseases. 2014;14(5):397–405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

