Association of RNA Biosignatures With Bacterial Infections in Febrile Infants Aged 60 Days or Younger
- PMID: 27552618
- PMCID: PMC5122927
- DOI: 10.1001/jama.2016.9207
Association of RNA Biosignatures With Bacterial Infections in Febrile Infants Aged 60 Days or Younger
Erratum in
-
Misidentification of Patients in the Abstract and Author Affiliations.JAMA. 2016 Nov 8;316(18):1924. doi: 10.1001/jama.2016.15459. JAMA. 2016. PMID: 27824993 No abstract available.
Abstract
Importance: Young febrile infants are at substantial risk of serious bacterial infections; however, the current culture-based diagnosis has limitations. Analysis of host expression patterns ("RNA biosignatures") in response to infections may provide an alternative diagnostic approach.
Objective: To assess whether RNA biosignatures can distinguish febrile infants aged 60 days or younger with and without serious bacterial infections.
Design, setting, and participants: Prospective observational study involving a convenience sample of febrile infants 60 days or younger evaluated for fever (temperature >38° C) in 22 emergency departments from December 2008 to December 2010 who underwent laboratory evaluations including blood cultures. A random sample of infants with and without bacterial infections was selected for RNA biosignature analysis. Afebrile healthy infants served as controls. Blood samples were collected for cultures and RNA biosignatures. Bioinformatics tools were applied to define RNA biosignatures to classify febrile infants by infection type.
Exposure: RNA biosignatures compared with cultures for discriminating febrile infants with and without bacterial infections and infants with bacteremia from those without bacterial infections.
Main outcomes and measures: Bacterial infection confirmed by culture. Performance of RNA biosignatures was compared with routine laboratory screening tests and Yale Observation Scale (YOS) scores.
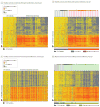
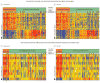
Results: Of 1883 febrile infants (median age, 37 days; 55.7% boys), RNA biosignatures were measured in 279 randomly selected infants (89 with bacterial infections-including 32 with bacteremia and 15 with urinary tract infections-and 190 without bacterial infections), and 19 afebrile healthy infants. Sixty-six classifier genes were identified that distinguished infants with and without bacterial infections in the test set with 87% (95% CI, 73%-95%) sensitivity and 89% (95% CI, 81%-93%) specificity. Ten classifier genes distinguished infants with bacteremia from those without bacterial infections in the test set with 94% (95% CI, 70%-100%) sensitivity and 95% (95% CI, 88%-98%) specificity. The incremental C statistic for the RNA biosignatures over the YOS score was 0.37 (95% CI, 0.30-0.43).
Conclusions and relevance: In this preliminary study, RNA biosignatures were defined to distinguish febrile infants aged 60 days or younger with vs without bacterial infections. Further research with larger populations is needed to refine and validate the estimates of test accuracy and to assess the clinical utility of RNA biosignatures in practice.
Conflict of interest statement
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Dr Mejias reports receiving personal fees from Abbvie, Novartis, and Janssen and grants from Gilead, Alios, and Janssen. Dr Ramilo reports receiving personal fees from HuMabs, Abbvie, Janssen, Medimmune, and Regeneron and grants from Janssen. All these fees and grants were not related to the current work. No other disclosures were reported.
Figures



Comment in
-
Genetics and the Evaluation of the Febrile Child.JAMA. 2016 Aug 23-30;316(8):824-5. doi: 10.1001/jama.2016.11137. JAMA. 2016. PMID: 27552615 No abstract available.
-
Diagnosis of Bacterial Infection Using a 2-Transcript Host RNA Signature in Febrile Infants 60 Days or Younger.JAMA. 2017 Apr 18;317(15):1577-1578. doi: 10.1001/jama.2017.1365. JAMA. 2017. PMID: 28418473 No abstract available.
References
-
- Byington CL, Rittichier KK, Bassett KE, et al. Serious bacterial infections in febrile infants younger than 90 days of age: the importance of ampicillin-resistant pathogens. Pediatrics. 2003;111(5 pt 1):964–968. - PubMed
-
- DeAngelis C, Joffe A, Wilson M, Willis E. Iatrogenic risks and financial costs of hospitalizing febrile infants. Am J Dis Child. 1983;137(12):1146–1149. - PubMed
-
- Cummings CA, Relman DA. Microbial forensics—“cross-examining pathogens. ” Science. 2002;296(5575):1976–1979. - PubMed
-
- Wilson ML. Clinically relevant, cost-effective clinical microbiology: strategies to decrease unnecessary testing. Am J Clin Pathol. 1997;107(2):154–167. - PubMed
-
- Zwang O, Albert RK. Analysis of strategies to improve cost-effectiveness of blood cultures. J Hosp Med. 2006;1(5):272–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

