CAZyChip: dynamic assessment of exploration of glycoside hydrolases in microbial ecosystems
- PMID: 27552843
- PMCID: PMC4994258
- DOI: 10.1186/s12864-016-2988-4
CAZyChip: dynamic assessment of exploration of glycoside hydrolases in microbial ecosystems
Abstract
Background: Microorganisms constitute a reservoir of enzymes involved in environmental carbon cycling and degradation of plant polysaccharides through their production of a vast variety of Glycoside Hydrolases (GH). The CAZyChip was developed to allow a rapid characterization at transcriptomic level of these GHs and to identify enzymes acting on hydrolysis of polysaccharides or glycans.
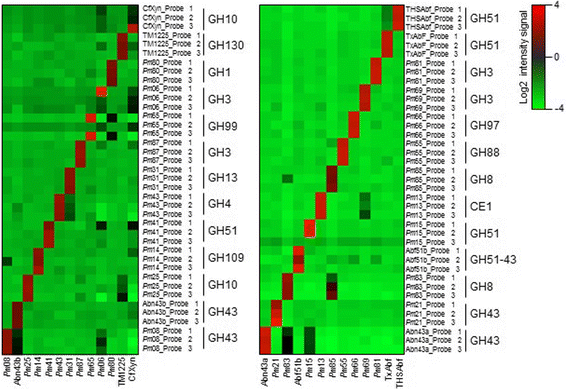
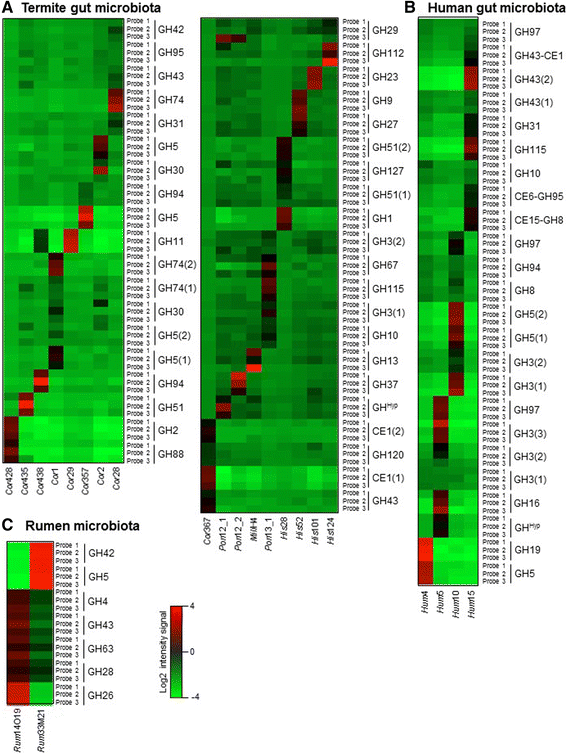
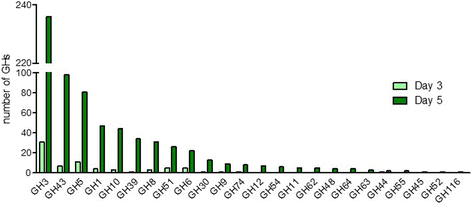
Results: This DNA biochip contains the signature of 55,220 bacterial GHs available in the CAZy database. Probes were designed using two softwares, and microarrays were directly synthesized using the in situ ink-jet technology. CAZyChip specificity and reproducibility was validated by hybridization of known GHs RNA extracted from recombinant E. coli strains, which were previously identified by a functional metagenomic approach. The GHs arsenal was also studied in bioprocess conditions using rumen derived microbiota.
Conclusions: The CAZyChip appears to be a user friendly tool for profiling the expression of a large variety of GHs. It can be used to study temporal variations of functional diversity, thereby facilitating the identification of new efficient candidates for enzymatic conversions from various ecosystems.
Keywords: CAZymes detection; Glycoside hydrolase; Microarray; Microbial functional diversity; Plant cell wall degradation; Transcriptomic analysis.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

