Nucleophosmin: from structure and function to disease development
- PMID: 27553022
- PMCID: PMC4995807
- DOI: 10.1186/s12867-016-0073-9
Nucleophosmin: from structure and function to disease development
Abstract
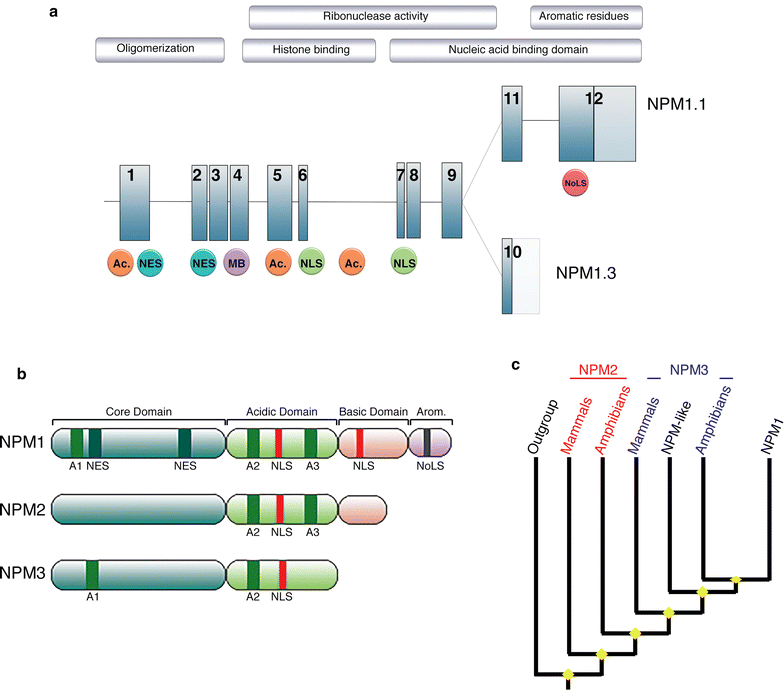
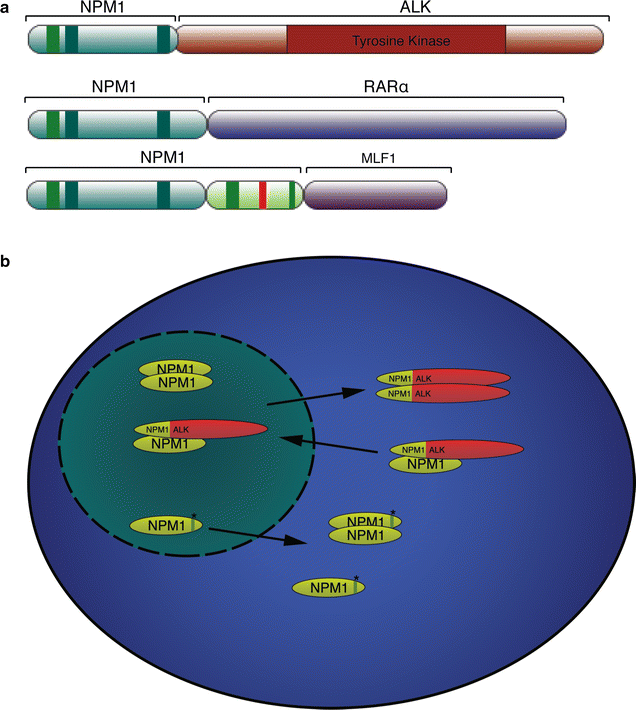
Nucleophosmin (NPM1) is a critical cellular protein that has been implicated in a number of pathways including mRNA transport, chromatin remodeling, apoptosis and genome stability. NPM1 function is a critical requirement for normal cellular biology as is underlined in cancer where NPM1 is commonly overexpressed, mutated, rearranged and sporadically deleted. Consistent with a multifunctional role within the cell, NPM1 can function not only as a proto-oncogene but also as a tumor suppressor. The aim of this review is to look at the less well-described role of NPM1 in the DNA repair pathways as well as the role of NPM1 in the regulation of apoptosis and its mutation in cancers.
Keywords: Apoptosis; Cancer; DNA repair; Nucleophosmin 1.
Figures




References
-
- Umekawa H, Chang JH, Correia JJ, Wang D, Wingfield PT, Olson MO. Nucleolar protein B23: bacterial expression, purification, oligomerization and secondary structures of two isoforms. Cell Mol Biol Res. 1993;39(7):635–645. - PubMed
-
- Dalenc F, Drouet J, Ader I, Delmas C, Rochaix P, Favre G, Cohen-Jonathan E, Toulas C. Increased expression of a COOH-truncated nucleophosmin resulting from alternative splicing is associated with cellular resistance to ionizing radiation in HeLa cells. Int J Cancer. 2002;100(6):662–668. doi: 10.1002/ijc.10558. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

