Stratification and therapeutic potential of PML in metastatic breast cancer
- PMID: 27553708
- PMCID: PMC4999521
- DOI: 10.1038/ncomms12595
Stratification and therapeutic potential of PML in metastatic breast cancer
Abstract
Patient stratification has been instrumental for the success of targeted therapies in breast cancer. However, the molecular basis of metastatic breast cancer and its therapeutic vulnerabilities remain poorly understood. Here we show that PML is a novel target in aggressive breast cancer. The acquisition of aggressiveness and metastatic features in breast tumours is accompanied by the elevated PML expression and enhanced sensitivity to its inhibition. Interestingly, we find that STAT3 is responsible, at least in part, for the transcriptional upregulation of PML in breast cancer. Moreover, PML targeting hampers breast cancer initiation and metastatic seeding. Mechanistically, this biological activity relies on the regulation of the stem cell gene SOX9 through interaction of PML with its promoter region. Altogether, we identify a novel pathway sustaining breast cancer aggressiveness that can be therapeutically exploited in combination with PML-based stratification.
Figures

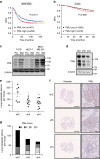
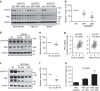



References
-
- Haber D. A., Gray N. S. & Baselga J. The evolving war on cancer. Cell 145, 19–24 (2011). - PubMed
-
- Normanno N. et al.. Prognostic applications of gene expression signatures in breast cancer. Oncology (Williston Park) 77, (Suppl 1): 2–8 (2009). - PubMed
-
- van de Vijver M. J. et al.. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

