Substrate Affinity Differentially Influences Protein Kinase C Regulation and Inhibitor Potency
- PMID: 27555323
- PMCID: PMC5063980
- DOI: 10.1074/jbc.M116.737601
Substrate Affinity Differentially Influences Protein Kinase C Regulation and Inhibitor Potency
Abstract
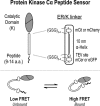
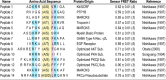
The overlapping network of kinase-substrate interactions provides exquisite specificity in cell signaling pathways, but also presents challenges to our ability to understand the mechanistic basis of biological processes. Efforts to dissect kinase-substrate interactions have been particularly limited by their inherently transient nature. Here, we use a library of FRET sensors to monitor these transient complexes, specifically examining weak interactions between the catalytic domain of protein kinase Cα and 14 substrate peptides. Combining results from this assay platform with those from standard kinase activity assays yields four novel insights into the kinase-substrate interaction. First, preferential binding of non-phosphorylated versus phosphorylated substrates leads to enhanced kinase-specific activity. Second, kinase-specific activity is inversely correlated with substrate binding affinity. Third, high affinity substrates can suppress phosphorylation of their low affinity counterparts. Finally, the substrate-competitive inhibitor bisindolylmaleimide I displaces low affinity substrates more potently leading to substrate selective inhibition of kinase activity. Overall, our approach complements existing structural and biophysical approaches to provide generalizable insights into the regulation of kinase activity.
Keywords: fluorescence resonance energy transfer (FRET); inhibitor; phosphorylation; protein kinase C (PKC); substrate specificity.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Pearce L. R., Komander D., and Alessi D. R. (2010) The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell. Biol. 11, 9–22 - PubMed
-
- Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 - PubMed
-
- de Oliveira P. S., Ferraz F. A., Pena D. A., Pramio D. T., Morais F. A., and Schechtman D. (2016) Revisiting protein kinase-substrate interactions: toward therapeutic development. Sci. Signal. 9, re3. - PubMed
-
- Hanks S. K., and Hunter T. (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

