A large-scale analysis of alternative splicing reveals a key role of QKI in lung cancer
- PMID: 27555542
- PMCID: PMC5423218
- DOI: 10.1016/j.molonc.2016.08.001
A large-scale analysis of alternative splicing reveals a key role of QKI in lung cancer
Abstract
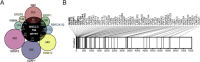
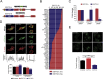
Increasing interest has been devoted in recent years to the understanding of alternative splicing in cancer. In this study, we performed a genome-wide analysis to identify cancer-associated splice variants in non-small cell lung cancer. We discovered and validated novel differences in the splicing of genes known to be relevant to lung cancer biology, such as NFIB, ENAH or SPAG9. Gene enrichment analyses revealed an important contribution of alternative splicing to cancer-related molecular functions, especially those involved in cytoskeletal dynamics. Interestingly, a substantial fraction of the altered genes found in our analysis were targets of the protein quaking (QKI), pointing to this factor as one of the most relevant regulators of alternative splicing in non-small cell lung cancer. We also found that ESYT2, one of the QKI targets, is involved in cytoskeletal organization. ESYT2-short variant inhibition in lung cancer cells resulted in a cortical distribution of actin whereas inhibition of the long variant caused an increase of endocytosis, suggesting that the cancer-associated splicing pattern of ESYT2 has a profound impact in the biology of cancer cells. Finally, we show that low nuclear QKI expression in non-small cell lung cancer is an independent prognostic factor for disease-free survival (HR = 2.47; 95% CI = 1.11-5.46, P = 0.026). In conclusion, we identified several splicing variants with functional relevance in lung cancer largely regulated by the splicing factor QKI, a tumor suppressor associated with prognosis in lung cancer.
Keywords: Alternative splicing; ESYT2; Non-small cell lung cancer; QKI.
Copyright © 2016 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Barretina, J. , Caponigro, G. , Stransky, N. , Venkatesan, K. , Margolin, A.A. , Kim, S. , Wilson, C.J. , Lehar, J. , Kryukov, G.V. , Sonkin, D. , Reddy, A. , Liu, M. , Murray, L. , Berger, M.F. , Monahan, J.E. , Morais, P. , Meltzer, J. , Korejwa, A. , Jane-Valbuena, J. , Mapa, F.A. , Thibault, J. , Bric-Furlong, E. , Raman, P. , Shipway, A. , Engels, I.H. , Cheng, J. , Yu, G.K. , Yu, J. , Aspesi, P. , de Silva, M. , Jagtap, K. , Jones, M.D. , Wang, L. , Hatton, C. , Palescandolo, E. , Gupta, S. , Mahan, S. , Sougnez, C. , Onofrio, R.C. , Liefeld, T. , MacConaill, L. , Winckler, W. , Reich, M. , Li, N. , Mesirov, J.P. , Gabriel, S.B. , Getz, G. , Ardlie, K. , Chan, V. , Myer, V.E. , Weber, B.L. , Porter, J. , Warmuth, M. , Finan, P. , Harris, J.L. , Meyerson, M. , Golub, T.R. , Morrissey, M.P. , Sellers, W.R. , Schlegel, R. , Garraway, L.A. , 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 483, 603–607. - PMC - PubMed
-
- Bechara, E.G. , Sebestyen, E. , Bernardis, I. , Eyras, E. , Valcarcel, J. , 2013. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cel. 52, 720–733. - PubMed
-
- Biamonti, G. , Catillo, M. , Pignataro, D. , Montecucco, A. , Ghigna, C. , 2014. The alternative splicing side of cancer. Semin. Cel. Dev. Biol. 32, 30–36. - PubMed
-
- Bian, Y. , Wang, L. , Lu, H. , Yang, G. , Zhang, Z. , Fu, H. , Lu, X. , Wei, M. , Sun, J. , Zhao, Q. , Dong, G. , Lu, Z. , 2012. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis. Biochem. Biophys. Res. Commun. 422, 187–193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

