Post-translational Serine/Threonine Phosphorylation and Lysine Acetylation: A Novel Regulatory Aspect of the Global Nitrogen Response Regulator GlnR in S. coelicolor M145
- PMID: 27556027
- PMCID: PMC4977719
- DOI: 10.3389/fmolb.2016.00038
Post-translational Serine/Threonine Phosphorylation and Lysine Acetylation: A Novel Regulatory Aspect of the Global Nitrogen Response Regulator GlnR in S. coelicolor M145
Abstract
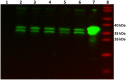
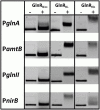
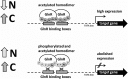
Soil-dwelling Streptomyces bacteria such as S.coelicolor have to constantly adapt to the nitrogen (N) availability in their habitat. Thus, strict transcriptional and post-translational control of the N-assimilation is fundamental for survival of this species. GlnR is a global response regulator that controls transcription of the genes related to the N-assimilation in S. coelicolor and other members of the Actinomycetales. GlnR represents an atypical orphan response regulator that is not activated by the phosphorylation of the conserved aspartate residue (Asp 50). We have applied transcriptional analysis, LC-MS/MS analysis and electrophoretic mobility shift assays (EMSAs) to understand the regulation of GlnR in S. coelicolor M145. The expression of glnR and GlnR-target genes was revisited under four different N-defined conditions and a complex N-rich condition. Although, the expression of selected GlnR-target genes was strongly responsive to changing N-concentrations, the glnR expression itself was independent of the N-availability. Using LC-MS/MSanalysis we demonstrated that GlnR was post-translationally modified. The post-translational modifications of GlnR comprise phosphorylation of the serine/threonine residues and acetylation of lysine residues. In the complex N-rich medium GlnR was phosphorylated on six serine/threonine residues and acetylated on one lysine residue. Under defined N-excess conditions only two phosphorylated residues were detected whereas under defined N-limiting conditions no phosphorylation was observed. GlnR phosphorylation is thus clearly correlated with N-rich conditions. Furthermore, GlnR was acetylated on four lysine residues independently of the N-concentration in the defined media and on only one lysine residue in the complex N-rich medium. Using EMSAs we demonstrated that phosphorylation inhibited the binding of GlnR to its targets genes, whereas acetylation had little influence on the formation of GlnR-DNA complex. This study clearly demonstrated that GlnR DNA-binding affinity is modulated by post-translational modifications in response to changing N-conditions in order to elicit a proper transcriptional response to the latter.
Keywords: GlnR; Streptomyces coelicolor; acetylation; nitrogen assimilation; phosphorylation; post-translational modifications; regulation.
Figures


References
-
- Amon J., Bräu T., Grimrath A., Hanssler E., Hasselt K., Höller M., et al. (2008). Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR. J. Bacteriol. 190, 7108–7116. 10.1128/JB.00855-08 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

