Production of Small Noncoding RNAs from the flamenco Locus Is Regulated by the gypsy Retrotransposon of Drosophila melanogaster
- PMID: 27558137
- PMCID: PMC5068851
- DOI: 10.1534/genetics.116.187922
Production of Small Noncoding RNAs from the flamenco Locus Is Regulated by the gypsy Retrotransposon of Drosophila melanogaster
Abstract
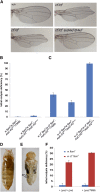
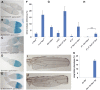
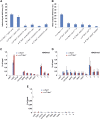
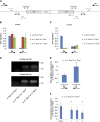
Protective mechanisms based on RNA silencing directed against the propagation of transposable elements are highly conserved in eukaryotes. The control of transposable elements is mediated by small noncoding RNAs, which derive from transposon-rich heterochromatic regions that function as small RNA-generating loci. These clusters are transcribed and the precursor transcripts are processed to generate Piwi-interacting RNAs (piRNAs) and endogenous small interfering RNAs (endo-siRNAs), which silence transposable elements in gonads and somatic tissues. The flamenco locus is a Drosophila melanogaster small RNA cluster that controls gypsy and other transposable elements, and has played an important role in understanding how small noncoding RNAs repress transposable elements. In this study, we describe a cosuppression mechanism triggered by new euchromatic gypsy insertions in genetic backgrounds carrying flamenco alleles defective in gypsy suppression. We found that the silencing of gypsy is accompanied by the silencing of other transposons regulated by flamenco, and of specific flamenco sequences from which small RNAs against gypsy originate. This cosuppression mechanism seems to depend on a post-transcriptional regulation that involves both endo-siRNA and piRNA pathways and is associated with the occurrence of developmental defects. In conclusion, we propose that new gypsy euchromatic insertions trigger a post-transcriptional silencing of gypsy sense and antisense sequences, which modifies the flamenco activity. This cosuppression mechanism interferes with some developmental processes, presumably by influencing the expression of specific genes.
Keywords: RNA silencing; ecdysis; primary transcript; small RNA; transposon.
Copyright © 2016 by the Genetics Society of America.
Figures


References
-
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., et al. , 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. - PubMed
-
- Bucheton A., 1995. The relationship between the flamenco gene and gypsy in Drosophila: how to tame a retrovirus. Trends Genet. 11: 349–353. - PubMed
-
- Buchon N., Vaury C., 2006. RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity 96: 195–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

