Real-Time Genome Sequencing of Resistant Bacteria Provides Precision Infection Control in an Institutional Setting
- PMID: 27558178
- PMCID: PMC5121374
- DOI: 10.1128/JCM.00790-16
Real-Time Genome Sequencing of Resistant Bacteria Provides Precision Infection Control in an Institutional Setting
Abstract
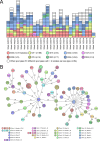
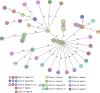
The increasing prevalence of multidrug-resistant (MDR) bacteria is a serious global challenge. Here, we studied prospectively whether bacterial whole-genome sequencing (WGS) for real-time MDR surveillance is technical feasible, returns actionable results, and is cost-beneficial. WGS was applied to all MDR isolates of four species (methicillin-resistant Staphylococcus aureus [MRSA], vancomycin-resistant Enterococcus faecium, MDR Escherichia coli, and MDR Pseudomonas aeruginosa) at the University Hospital Muenster, Muenster, Germany, a tertiary care hospital with 1,450 beds, during two 6-month intervals. Turnaround times (TAT) were measured, and total costs for sequencing per isolate were calculated. After cancelling prior policies of preemptive isolation of patients harboring certain Gram-negative MDR bacteria in risk areas, the second interval was conducted. During interval I, 645 bacterial isolates were sequenced. From culture, TATs ranged from 4.4 to 5.3 days, and costs were €202.49 per isolate. During interval II, 550 bacterial isolates were sequenced. Hospital-wide transmission rates of the two most common species (MRSA and MDR E. coli) were low during interval I (5.8% and 2.3%, respectively) and interval II (4.3% and 5.0%, respectively). Cancellation of isolation of patients infected with non-pan-resistant MDR E. coli in risk wards did not increase transmission. Comparing sequencing costs with avoided costs mostly due to fewer blocked beds during interval II, we saved in excess of €200,000. Real-time microbial WGS in our institution was feasible, produced precise actionable results, helped us to monitor transmission rates that remained low following a modification in isolation procedures, and ultimately saved costs.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf Accessed 2 January 2015.
-
- Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi:10.1371/journal.pone.0022751. - DOI - PMC - PubMed
-
- Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J, Xi F, Li S, Li Y, Zhang Z, Yang X, Zhao M, Wang P, Guan Y, Cen Z, Zhao X, Christner M, Kobbe R, Loos S, Oh J, Yang L, Danchin A, Gao GF, Song Y, Li Y, Yang H, Wang J, Xu J, Pallen MJ, Wang J, Aepfelbacher M, Yang R; E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium. 2011. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med 365:718–724. doi:10.1056/NEJMoa1107643. - DOI - PubMed
-
- Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, Ip CL, Wilson DJ, Didelot X, O'Connor L, Lay R, Buck D, Kearns AM, Shaw A, Paul J, Wilcox MH, Donnelly PJ, Peto TE, Walker AS, Crook DW. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:e001124. doi:10.1136/bmjopen-2012-001124. - DOI - PMC - PubMed
-
- Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi:10.1126/science.1182395. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

