Regulatory Role of Circular RNAs and Neurological Disorders
- PMID: 27558238
- PMCID: PMC5955391
- DOI: 10.1007/s12035-016-0055-4
Regulatory Role of Circular RNAs and Neurological Disorders
Abstract
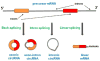
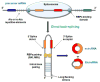
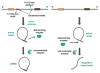
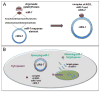
Circular RNAs (circRNAs) are a class of long noncoding RNAs that are characterized by the presence of covalently linked ends and have been found in all life kingdoms. Exciting studies in regulatory roles of circRNAs are emerging. Here, we summarize classification, characteristics, biogenesis, and regulatory functions of circRNAs. CircRNAs are found to be preferentially expressed along neural genes and in neural tissues. We thus highlight the association of circRNA dysregulation with neurodegenerative diseases such as Alzheimer's disease. Investigation of regulatory role of circRNAs will shed novel light in gene expression mechanisms during development and under disease conditions and may identify circRNAs as new biomarkers for aging and neurodegenerative disorders.
Keywords: Back-splicing; Circular RNA; Neurodegeneration; Regulatory RNA; microRNA sponge.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

