A conserved role of the RSC chromatin remodeler in the establishment of nucleosome-depleted regions
- PMID: 27558480
- PMCID: PMC5383693
- DOI: 10.1007/s00294-016-0642-y
A conserved role of the RSC chromatin remodeler in the establishment of nucleosome-depleted regions
Abstract
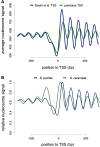
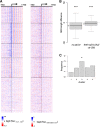
The occupancy of nucleosomes governs access to the eukaryotic genomes and results from a combination of biophysical features and the effect of ATP-dependent remodelling complexes. Most promoter regions show a conserved pattern characterized by a nucleosome-depleted region (NDR) flanked by nucleosomal arrays. The conserved RSC remodeler was reported to be critical to establish NDR in vivo in budding yeast but other evidences suggested that this activity may not be conserved in fission yeast. By reanalysing and expanding previously published data, we propose that NDR formation requires, at least partially, RSC in both yeast species. We also discuss the most prominent biological role of RSC and the possibility that non-essential subunits do not define alternate versions of the complex.
Keywords: Chromatin; Mitosis; Nucleosome; RSC; Yeast.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures

References
-
- Anuchin AM, Goncharenko AV, Demidenok OI, Kaprel’iants AS. Histone-like proteins of bacteria (review) Prikl Biokhim Mikrobiol. 2011;47:635–641. - PubMed
-
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

