Diminishing-returns epistasis among random beneficial mutations in a multicellular fungus
- PMID: 27559062
- PMCID: PMC5013798
- DOI: 10.1098/rspb.2016.1376
Diminishing-returns epistasis among random beneficial mutations in a multicellular fungus
Abstract
Adaptive evolution ultimately is fuelled by mutations generating novel genetic variation. Non-additivity of fitness effects of mutations (called epistasis) may affect the dynamics and repeatability of adaptation. However, understanding the importance and implications of epistasis is hampered by the observation of substantial variation in patterns of epistasis across empirical studies. Interestingly, some recent studies report increasingly smaller benefits of beneficial mutations once genotypes become better adapted (called diminishing-returns epistasis) in unicellular microbes and single genes. Here, we use Fisher's geometric model (FGM) to generate analytical predictions about the relationship between the effect size of mutations and the extent of epistasis. We then test these predictions using the multicellular fungus Aspergillus nidulans by generating a collection of 108 strains in either a poor or a rich nutrient environment that each carry a beneficial mutation and constructing pairwise combinations using sexual crosses. Our results support the predictions from FGM and indicate negative epistasis among beneficial mutations in both environments, which scale with mutational effect size. Hence, our findings show the importance of diminishing-returns epistasis among beneficial mutations also for a multicellular organism, and suggest that this pattern reflects a generic constraint operating at diverse levels of biological organization.
Keywords: Aspergillus nidulans; Fisher's geometric model; adaptation; beneficial mutation; epistasis.
© 2016 The Author(s).
Figures
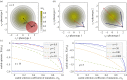
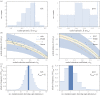
 , respectively. The straight grey lines show the relationship ɛ = −s.
, respectively. The straight grey lines show the relationship ɛ = −s.
References
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
