Total HIV-1 DNA, a Marker of Viral Reservoir Dynamics with Clinical Implications
- PMID: 27559075
- PMCID: PMC5010749
- DOI: 10.1128/CMR.00015-16
Total HIV-1 DNA, a Marker of Viral Reservoir Dynamics with Clinical Implications
Abstract
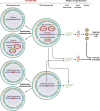
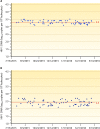
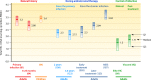
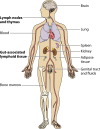
HIV-1 DNA persists in infected cells despite combined antiretroviral therapy (cART), forming viral reservoirs. Recent trials of strategies targeting latent HIV reservoirs have rekindled hopes of curing HIV infection, and reliable markers are thus needed to evaluate viral reservoirs. Total HIV DNA quantification is simple, standardized, sensitive, and reproducible. Total HIV DNA load influences the course of the infection and is therefore clinically relevant. In particular, it is predictive of progression to AIDS and death, independently of HIV RNA load and the CD4 cell count. Baseline total HIV DNA load is predictive of the response to cART. It declines during cART but remains quantifiable, at a level that reflects both the history of infection (HIV RNA zenith, CD4 cell count nadir) and treatment efficacy (residual viremia, cumulative viremia, immune restoration, immune cell activation). Total HIV DNA load in blood is also predictive of the presence and severity of some HIV-1-associated end-organ disorders. It can be useful to guide individual treatment, notably, therapeutic de-escalation. Although it does not distinguish between replication-competent and -defective latent viruses, the total HIV DNA load in blood, tissues, and cells provides insights into HIV pathogenesis, probably because all viral forms participate in host cell activation and HIV pathogenesis. Total HIV DNA is thus a biomarker of HIV reservoirs, which can be defined as all infected cells and tissues containing all forms of HIV persistence that participate in pathogenesis. This participation may occur through the production of new virions, creating new cycles of infection and disseminating infected cells; maintenance or amplification of reservoirs by homeostatic cell proliferation; and viral transcription and synthesis of viral proteins without new virion production. These proteins can induce immune activation, thus participating in the vicious circle of HIV pathogenesis.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures








References
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi:10.1038/8394. - DOI - PubMed
-
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi:10.1038/387183a0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

