C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling
- PMID: 27559131
- PMCID: PMC5063613
- DOI: 10.1091/mbc.E16-01-0003
C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling
Abstract
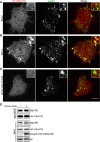
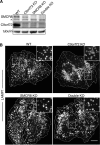
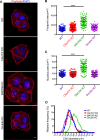
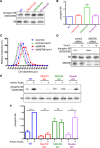
Hexanucleotide expansion in an intron of the C9orf72 gene causes amyotrophic lateral sclerosis and frontotemporal dementia. However, beyond bioinformatics predictions that suggested structural similarity to folliculin, the Birt-Hogg-Dubé syndrome tumor suppressor, little is known about the normal functions of the C9orf72 protein. To address this problem, we used genome-editing strategies to investigate C9orf72 interactions, subcellular localization, and knockout (KO) phenotypes. We found that C9orf72 robustly interacts with SMCR8 (a protein of previously unknown function). We also observed that C9orf72 localizes to lysosomes and that such localization is negatively regulated by amino acid availability. Analysis of C9orf72 KO, SMCR8 KO, and double-KO cell lines revealed phenotypes that are consistent with a function for C9orf72 at lysosomes. These include abnormally swollen lysosomes in the absence of C9orf72 and impaired responses of mTORC1 signaling to changes in amino acid availability (a lysosome-dependent process) after depletion of either C9orf72 or SMCR8. Collectively these results identify strong physical and functional interactions between C9orf72 and SMCR8 and support a lysosomal site of action for this protein complex.
© 2016 Amick et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures


References
-
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, 3rd, Hartley JL, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci USA. 2006;103:15552–15557. - PMC - PubMed
-
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

