Cyclisation mechanisms in the biosynthesis of ribosomally synthesised and post-translationally modified peptides
- PMID: 27559376
- PMCID: PMC4979651
- DOI: 10.3762/bjoc.12.120
Cyclisation mechanisms in the biosynthesis of ribosomally synthesised and post-translationally modified peptides
Abstract
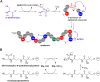
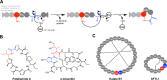
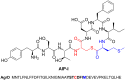
Ribosomally synthesised and post-translationally modified peptides (RiPPs) are a large class of natural products that are remarkably chemically diverse given an intrinsic requirement to be assembled from proteinogenic amino acids. The vast chemical space occupied by RiPPs means that they possess a wide variety of biological activities, and the class includes antibiotics, co-factors, signalling molecules, anticancer and anti-HIV compounds, and toxins. A considerable amount of RiPP chemical diversity is generated from cyclisation reactions, and the current mechanistic understanding of these reactions will be discussed here. These cyclisations involve a diverse array of chemical reactions, including 1,4-nucleophilic additions, [4 + 2] cycloadditions, ATP-dependent heterocyclisation to form thiazolines or oxazolines, and radical-mediated reactions between unactivated carbons. Future prospects for RiPP pathway discovery and characterisation will also be highlighted.
Keywords: RiPPs; biosynthesis; cyclisation; enzymes; peptides.
Figures








References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
