Application of Cu(I)-catalyzed azide-alkyne cycloaddition for the design and synthesis of sequence specific probes targeting double-stranded DNA
- PMID: 27559384
- PMCID: PMC4979877
- DOI: 10.3762/bjoc.12.128
Application of Cu(I)-catalyzed azide-alkyne cycloaddition for the design and synthesis of sequence specific probes targeting double-stranded DNA
Abstract
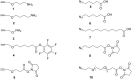
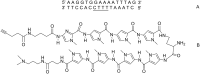
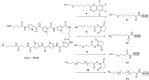
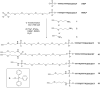
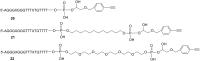
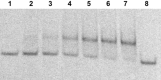
Efficient protocols based on Cu(I)-catalyzed azide-alkyne cycloaddition were developed for the synthesis of conjugates of pyrrole-imidazole polyamide minor groove binders (MGB) with fluorophores and with triplex-forming oligonucleotides (TFOs). Diverse bifunctional linkers were synthesized and used for the insertion of terminal azides or alkynes into TFOs and MGBs. The formation of stable triple helices by TFO-MGB conjugates was evaluated by gel-shift experiments. The presence of MGB in these conjugates did not affect the binding parameters (affinity and triplex stability) of the parent TFOs.
Keywords: Cu(I)-catalyzed azide–alkyne cycloaddition; binding affinity; click chemistry; pyrrole–imidazole polyamides; sequence specificity: DNA; triplex-forming oligonucleotides.
Figures
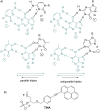
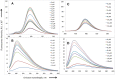
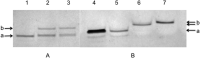
 ” and “
” and “ ”. The duplex is in green, TFO is in black. B) Chemical structure of the TINA molecule inserted in the TFO structure of the DNA triplex as a bulge.
”. The duplex is in green, TFO is in black. B) Chemical structure of the TINA molecule inserted in the TFO structure of the DNA triplex as a bulge.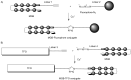

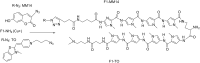


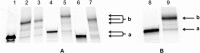

Similar articles
-
Head-to-head bis-hairpin polyamide minor groove binders and their conjugates with triplex-forming oligonucleotides: studies of interaction with target double-stranded DNA.J Biomol Struct Dyn. 2007 Aug;25(1):61-76. doi: 10.1080/07391102.2007.10507156. J Biomol Struct Dyn. 2007. PMID: 17676939
-
Synthesis of C-5, C-2' and C-4'-neomycin-conjugated triplex forming oligonucleotides and their affinity to DNA-duplexes.Bioorg Med Chem. 2015 Aug 1;23(15):4472-4480. doi: 10.1016/j.bmc.2015.06.013. Epub 2015 Jun 14. Bioorg Med Chem. 2015. PMID: 26118338
-
Sequence-specific conjugates of oligo(2'-O-methylribonucleotides) and hairpin oligocarboxamide minor-groove binders: design, synthesis, and binding studies with double-stranded DNA.Chem Biodivers. 2005 Jul;2(7):936-52. doi: 10.1002/cbdv.200590071. Chem Biodivers. 2005. PMID: 17193185
-
Electrophilic Azides for Materials Synthesis and Chemical Biology.Acc Chem Res. 2020 Apr 21;53(4):937-948. doi: 10.1021/acs.accounts.0c00046. Epub 2020 Mar 24. Acc Chem Res. 2020. PMID: 32207916 Review.
-
Triplex-forming oligonucleotides as an anti-gene technique for cancer therapy.Front Pharmacol. 2022 Dec 21;13:1007723. doi: 10.3389/fphar.2022.1007723. eCollection 2022. Front Pharmacol. 2022. PMID: 36618947 Free PMC article. Review.
Cited by
-
Recent Advances in Nucleic Acid Targeting Probes and Supramolecular Constructs Based on Pyrene-Modified Oligonucleotides.Molecules. 2017 Nov 30;22(12):2108. doi: 10.3390/molecules22122108. Molecules. 2017. PMID: 29189716 Free PMC article. Review.
-
Optimizing Mannose "Click" Conjugation to Polymeric Nanoparticles for Targeted siRNA Delivery to Human and Murine Macrophages.ACS Omega. 2019 Oct 1;4(16):16756-16767. doi: 10.1021/acsomega.9b01465. eCollection 2019 Oct 15. ACS Omega. 2019. PMID: 31646220 Free PMC article.
References
-
- Escudé C, Sun J-S. DNA Major Groove Binders: Triple Helix-Forming Oligonucleotides, Triple Helix-Specific DNA Ligands and Cleaving Agents. In: Waring M J, Chaires J B, editors. DNA Binders and Related Subjects. Vol. 253. Berlin, Germany: Springer; 2005. pp. 109–148. ((Topics in Current Chemistry)). - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
