Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells
- PMID: 27560004
- PMCID: PMC5056783
- DOI: 10.1080/15476286.2016.1218588
Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells
Abstract
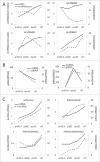
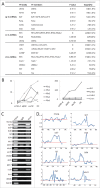
Among all tissues of the metazoa, the transcritpome of testis displays the highest diversity and specificity. However, its composition and dynamics during spermatogenesis have not been fully understood. Here, we have identified 20,639 message RNAs (mRNAs), 7,168 long non-coding RNAs (lncRNAs) and 15,101 circular RNAs (circRNAs) in mouse spermatogenic cells, and found many of them were specifically expressed in testes. lncRNAs are significantly more testis-specific than mRNAs. At all stages, mRNAs are generally more abundant than lncRNAs, and linear transcripts are more abundant than circRNAs. We showed that the productions of circRNAs and piRNAs were highly regulated instead of random processes. Based on the results of a small-scale functional screening experiment using cultured mouse spermatogonial stem cells, many evolutionarily conserved lncRNAs are likely to play roles in spermatogenesis. Typical classes of transcription factor binding sites are enriched in the promoters of testis-specific m/lncRNA genes. Target genes of CREM and RFX2, 2 key TFs for spermatogenesis, were further validated by using ChIP-chip assays and RNA-seq on RFX2-knockout spermatogenic cells. Our results contribute to the current understanding of the transcriptomic complexity of spermatogenic cells and provide a valuable resource from which many candidate genes may be selected for further functional studies.
Keywords: circRNAs; expression; lncRNAs; mRNAs; spermatogenesis; testis; transcription.
Figures



References
-
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet 2014; 15:423-37; PMID:24776770; http://dx.doi.org/ 10.1038/nrg3722 - DOI - PMC - PubMed
-
- Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Computational Biol 2009; 5:e1000598; PMID:20011106; http://dx.doi.org/ 10.1371/journal.pcbi.1000598 - DOI - PMC - PubMed
-
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, et al.. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 2013; 3:2179-90; PMID:23791531; http://dx.doi.org/ 10.1016/j.celrep.2013.05.031 - DOI - PubMed
-
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A 2003; 100:12201-6; PMID:14526100; http://dx.doi.org/ 10.1073/pnas.1635054100 - DOI - PMC - PubMed
-
- Gardini A, Shiekhattar R. The many faces of long noncoding RNAs. FEBS J 2015; 282:1647-57; PMID:25303371; http://dx.doi.org/ 10.1111/febs.13101 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
