DockQ: A Quality Measure for Protein-Protein Docking Models
- PMID: 27560519
- PMCID: PMC4999177
- DOI: 10.1371/journal.pone.0161879
DockQ: A Quality Measure for Protein-Protein Docking Models
Abstract
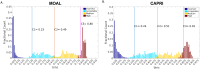
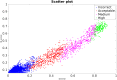
The state-of-the-art to assess the structural quality of docking models is currently based on three related yet independent quality measures: Fnat, LRMS, and iRMS as proposed and standardized by CAPRI. These quality measures quantify different aspects of the quality of a particular docking model and need to be viewed together to reveal the true quality, e.g. a model with relatively poor LRMS (>10Å) might still qualify as 'acceptable' with a descent Fnat (>0.50) and iRMS (<3.0Å). This is also the reason why the so called CAPRI criteria for assessing the quality of docking models is defined by applying various ad-hoc cutoffs on these measures to classify a docking model into the four classes: Incorrect, Acceptable, Medium, or High quality. This classification has been useful in CAPRI, but since models are grouped in only four bins it is also rather limiting, making it difficult to rank models, correlate with scoring functions or use it as target function in machine learning algorithms. Here, we present DockQ, a continuous protein-protein docking model quality measure derived by combining Fnat, LRMS, and iRMS to a single score in the range [0, 1] that can be used to assess the quality of protein docking models. By using DockQ on CAPRI models it is possible to almost completely reproduce the original CAPRI classification into Incorrect, Acceptable, Medium and High quality. An average PPV of 94% at 90% Recall demonstrating that there is no need to apply predefined ad-hoc cutoffs to classify docking models. Since DockQ recapitulates the CAPRI classification almost perfectly, it can be viewed as a higher resolution version of the CAPRI classification, making it possible to estimate model quality in a more quantitative way using Z-scores or sum of top ranked models, which has been so valuable for the CASP community. The possibility to directly correlate a quality measure to a scoring function has been crucial for the development of scoring functions for protein structure prediction, and DockQ should be useful in a similar development in the protein docking field. DockQ is available at http://github.com/bjornwallner/DockQ/.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- The Protein Structure Initiative: achievements and visions for the future [Internet]. [cited 15 Jun 2016]. Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3318194/ - PMC - PubMed
-
- 3did: a catalog of domain-based interactions of known three-dimensional structure [Internet]. [cited 15 Jun 2016]. Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3965002/ - PMC - PubMed
-
- Zemla A, Venclovas C, Moult J, Fidelis K. Processing and analysis of CASP3 protein structure predictions. Proteins. 1999;Suppl 3: 22–29. - PubMed
-
- Siew N, Elofsson A, Rychlewski L, Fischer D. MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics. 2000;16: 776–785. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

