Insufficient Evidence for Rare Activation of Latent HIV in the Absence of Reservoir-Reducing Interventions
- PMID: 27560936
- PMCID: PMC4999146
- DOI: 10.1371/journal.ppat.1005679
Insufficient Evidence for Rare Activation of Latent HIV in the Absence of Reservoir-Reducing Interventions
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
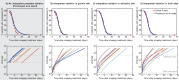
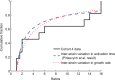
Comment in
-
Modeling of Experimental Data Supports HIV Reactivation from Latency after Treatment Interruption on Average Once Every 5-8 Days.PLoS Pathog. 2016 Aug 25;12(8):e1005740. doi: 10.1371/journal.ppat.1005740. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27560972 Free PMC article. No abstract available.
Comment on
-
Standing genetic variation and the evolution of drug resistance in HIV.PLoS Comput Biol. 2012;8(6):e1002527. doi: 10.1371/journal.pcbi.1002527. Epub 2012 Jun 7. PLoS Comput Biol. 2012. PMID: 22685388 Free PMC article.
Similar articles
-
Reversal of Latency as Part of a Cure for HIV-1.Trends Microbiol. 2016 Feb;24(2):90-97. doi: 10.1016/j.tim.2015.11.003. Epub 2015 Dec 11. Trends Microbiol. 2016. PMID: 26690612 Review.
-
Humanized Mouse Model of HIV-1 Latency with Enrichment of Latent Virus in PD-1+ and TIGIT+ CD4 T Cells.J Virol. 2019 May 1;93(10):e02086-18. doi: 10.1128/JVI.02086-18. Print 2019 May 15. J Virol. 2019. PMID: 30842333 Free PMC article.
-
Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure.J Virol. 2002 Sep;76(18):9481-92. doi: 10.1128/jvi.76.18.9481-9492.2002. J Virol. 2002. PMID: 12186930 Free PMC article.
-
Viral and latent reservoir persistence in HIV-1-infected patients on therapy.PLoS Comput Biol. 2006 Oct;2(10):e135. doi: 10.1371/journal.pcbi.0020135. Epub 2006 Aug 28. PLoS Comput Biol. 2006. PMID: 17040122 Free PMC article.
-
The latent HIV-1 reservoir in patients undergoing HAART: an archive of pre-HAART drug resistance.J Antimicrob Chemother. 2005 Apr;55(4):410-2. doi: 10.1093/jac/dki038. Epub 2005 Feb 22. J Antimicrob Chemother. 2005. PMID: 15728140 Review.
Cited by
-
Viral diversity is an obligate consideration in CRISPR/Cas9 designs for targeting the HIV reservoir.BMC Biol. 2018 Jul 11;16(1):75. doi: 10.1186/s12915-018-0544-1. BMC Biol. 2018. PMID: 29996827 Free PMC article.
-
Modeling of Experimental Data Supports HIV Reactivation from Latency after Treatment Interruption on Average Once Every 5-8 Days.PLoS Pathog. 2016 Aug 25;12(8):e1005740. doi: 10.1371/journal.ppat.1005740. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27560972 Free PMC article. No abstract available.
-
Estimating Initial Viral Levels during Simian Immunodeficiency Virus/Human Immunodeficiency Virus Reactivation from Latency.J Virol. 2018 Jan 2;92(2):e01667-17. doi: 10.1128/JVI.01667-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118123 Free PMC article.
-
Unified model of short- and long-term HIV viral rebound for clinical trial planning.J R Soc Interface. 2021 Apr;18(177):20201015. doi: 10.1098/rsif.2020.1015. Epub 2021 Apr 14. J R Soc Interface. 2021. PMID: 33849338 Free PMC article.
-
Dynamics of latent HIV under clonal expansion.PLoS Pathog. 2021 Dec 20;17(12):e1010165. doi: 10.1371/journal.ppat.1010165. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34929000 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- T15LM007079 /NIH/International
- P30AI060354/NIH/International
- P30 AI094189/AI/NIAID NIH HHS/United States
- U19 AI096109/AI/NIAID NIH HHS/United States
- R01AI043222 /NIH/International
- U19AI096109 /NIH/International
- R01 AI043222/AI/NIAID NIH HHS/United States
- U19 AI096113/AI/NIAID NIH HHS/United States
- AI096113 /NIH/International
- P30AI094189/NIH/International
- T15 LM007079/LM/NLM NIH HHS/United States
- DP5 OD019851/OD/NIH HHS/United States
- R01 GM117591/GM/NIGMS NIH HHS/United States
- R01GM117591 /NIH /International
- P30 AI060354/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

