Tissue- and Time-Specific Expression of Otherwise Identical tRNA Genes
- PMID: 27560950
- PMCID: PMC4999229
- DOI: 10.1371/journal.pgen.1006264
Tissue- and Time-Specific Expression of Otherwise Identical tRNA Genes
Abstract
Codon usage bias affects protein translation because tRNAs that recognize synonymous codons differ in their abundance. Although the current dogma states that tRNA expression is exclusively regulated by intrinsic control elements (A- and B-box sequences), we revealed, using a reporter that monitors the levels of individual tRNA genes in Caenorhabditis elegans, that eight tryptophan tRNA genes, 100% identical in sequence, are expressed in different tissues and change their expression dynamically. Furthermore, the expression levels of the sup-7 tRNA gene at day 6 were found to predict the animal's lifespan. We discovered that the expression of tRNAs that reside within introns of protein-coding genes is affected by the host gene's promoter. Pairing between specific Pol II genes and the tRNAs that are contained in their introns is most likely adaptive, since a genome-wide analysis revealed that the presence of specific intronic tRNAs within specific orthologous genes is conserved across Caenorhabditis species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




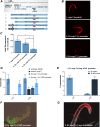
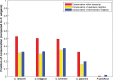
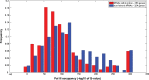
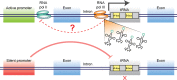
Similar articles
-
Evidence of multifaceted functions of codon usage in translation within the model beetle Tribolium castaneum.DNA Res. 2019 Dec 1;26(6):473-484. doi: 10.1093/dnares/dsz025. DNA Res. 2019. PMID: 31922535 Free PMC article.
-
Molecular Coping Mechanisms: Reprogramming tRNAs To Regulate Codon-Biased Translation of Stress Response Proteins.Acc Chem Res. 2023 Dec 5;56(23):3504-3514. doi: 10.1021/acs.accounts.3c00572. Epub 2023 Nov 22. Acc Chem Res. 2023. PMID: 37992267 Free PMC article.
-
Clustering of codons with rare cognate tRNAs in human genes suggests an extra level of expression regulation.PLoS Genet. 2009 Jul;5(7):e1000548. doi: 10.1371/journal.pgen.1000548. Epub 2009 Jul 3. PLoS Genet. 2009. PMID: 19578405 Free PMC article.
-
tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes.Trends Genet. 2000 Jul;16(7):287-9. doi: 10.1016/s0168-9525(00)02041-2. Trends Genet. 2000. PMID: 10858656 Review. No abstract available.
-
Synonymous Codons: Choose Wisely for Expression.Trends Genet. 2017 Apr;33(4):283-297. doi: 10.1016/j.tig.2017.02.001. Epub 2017 Mar 12. Trends Genet. 2017. PMID: 28292534 Free PMC article. Review.
Cited by
-
Expression of the Neuronal tRNA n-Tr20 Regulates Synaptic Transmission and Seizure Susceptibility.Neuron. 2020 Oct 14;108(1):193-208.e9. doi: 10.1016/j.neuron.2020.07.023. Epub 2020 Aug 26. Neuron. 2020. PMID: 32853550 Free PMC article.
-
ALL-tRNAseq enables robust tRNA profiling in tissue samples.Genes Dev. 2023 Mar 1;37(5-6):243-257. doi: 10.1101/gad.350233.122. Epub 2023 Feb 21. Genes Dev. 2023. PMID: 36810209 Free PMC article.
-
Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon-Codon Use.Biomolecules. 2017 Mar 8;7(1):26. doi: 10.3390/biom7010026. Biomolecules. 2017. PMID: 28282871 Free PMC article. Review.
-
On the Track of the Missing tRNA Genes: A Source of Non-Canonical Functions?Front Mol Biosci. 2021 Mar 16;8:643701. doi: 10.3389/fmolb.2021.643701. eCollection 2021. Front Mol Biosci. 2021. PMID: 33796548 Free PMC article. Review.
-
The stop-and-go traffic regulating protein biogenesis: How translation kinetics controls proteostasis.J Biol Chem. 2019 Feb 8;294(6):2076-2084. doi: 10.1074/jbc.REV118.002814. Epub 2018 Nov 30. J Biol Chem. 2019. PMID: 30504455 Free PMC article. Review.
References
-
- Crombie T, Swaffield JC, Brown AJ. Protein folding within the cell is influenced by controlled rates of polypeptide elongation. J Mol Biol. 1992;228: 7–12. Available: http://www.ncbi.nlm.nih.gov/pubmed/1447795 - PubMed
-
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984;3: 2895–8. Available: http://www.ncbi.nlm.nih.gov/pubmed/6396082 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

