Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status
- PMID: 27561551
- PMCID: PMC5007461
- DOI: 10.1038/ncomms12645
Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status
Abstract
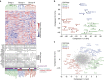
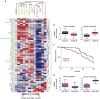
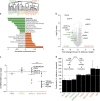
A cell line representative of human high-grade serous ovarian cancer (HGSOC) should not only resemble its tumour of origin at the molecular level, but also demonstrate functional utility in pre-clinical investigations. Here, we report the integrated proteomic analysis of 26 ovarian cancer cell lines, HGSOC tumours, immortalized ovarian surface epithelial cells and fallopian tube epithelial cells via a single-run mass spectrometric workflow. The in-depth quantification of >10,000 proteins results in three distinct cell line categories: epithelial (group I), clear cell (group II) and mesenchymal (group III). We identify a 67-protein cell line signature, which separates our entire proteomic data set, as well as a confirmatory publicly available CPTAC/TCGA tumour proteome data set, into a predominantly epithelial and mesenchymal HGSOC tumour cluster. This proteomics-based epithelial/mesenchymal stratification of cell lines and human tumours indicates a possible origin of HGSOC either from the fallopian tube or from the ovarian surface epithelium.
Figures



References
-
- Koonings P. P., Campbell K., Mishell D. R. Jr & Grimes D. A. Relative frequency of primary ovarian neoplasms: a 10-year review. Obstet. Gynecol. 74, 921–926 (1989). - PubMed
-
- Callahan M. J. et al.. Primary fallopian tube malignancies in BRCA-positive women undergoing surgery for ovarian cancer risk reduction. J. Clin. Oncol. 25, 3985–3990 (2007). - PubMed
-
- Kindelberger D. W. et al.. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: evidence for a causal relationship. Am. J. Surg. Pathol. 31, 161–169 (2007). - PubMed
-
- Piek J. M. et al.. Dysplastic changes in prophylactically removed fallopian tubes of women predisposed to developing ovarian cancer. J. Pathol. 195, 451–456 (2001). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

