Software-based analysis of bacteriophage genomes, physical ends, and packaging strategies
- PMID: 27561606
- PMCID: PMC5000459
- DOI: 10.1186/s12864-016-3018-2
Software-based analysis of bacteriophage genomes, physical ends, and packaging strategies
Abstract
Background: Phage genome analysis is a rapidly growing field. Recurrent obstacles include software access and usability, as well as genome sequences that vary in sequence orientation and/or start position. Here we describe modifications to the phage comparative genomics software program, Phamerator, provide public access to the code, and include instructions for creating custom Phamerator databases. We further report genomic analysis techniques to determine phage packaging strategies and identification of the physical ends of phage genomes.
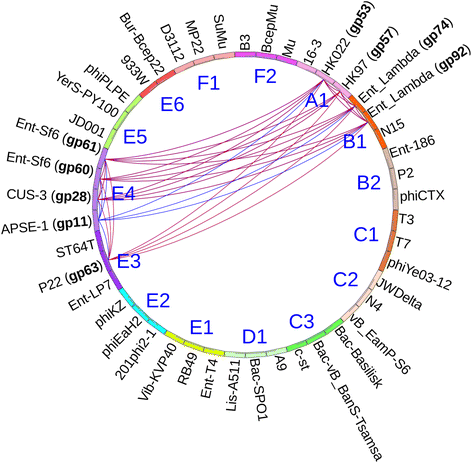
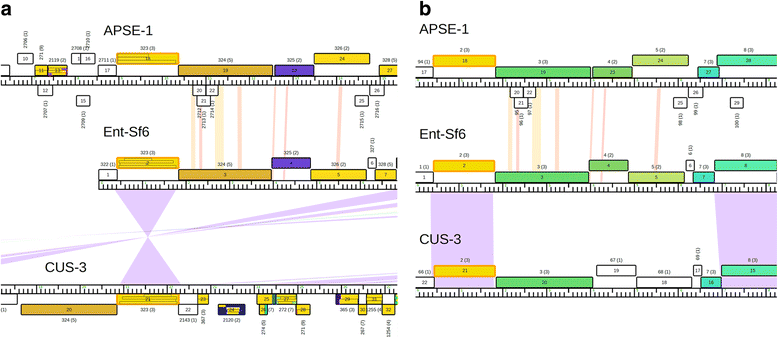
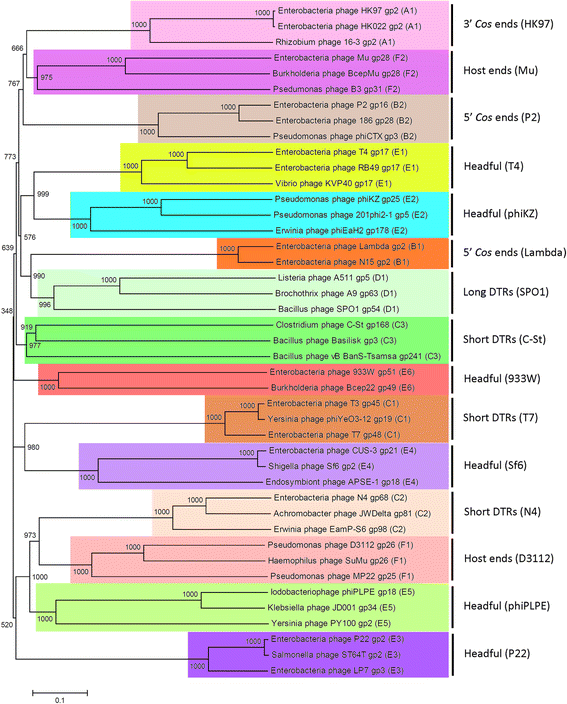
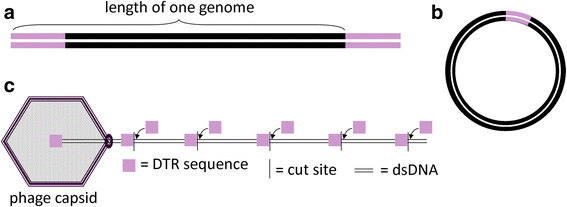
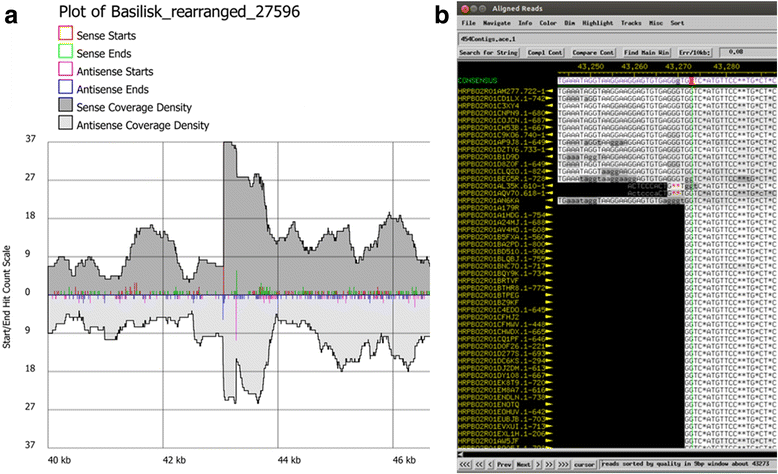
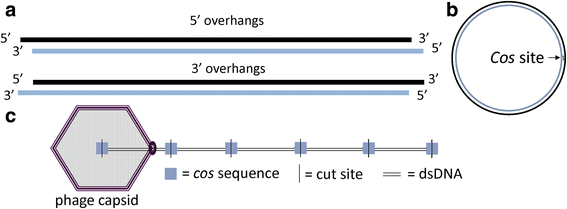
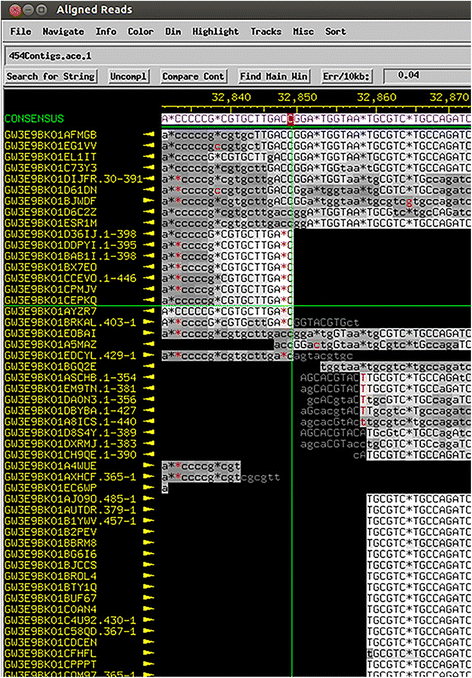
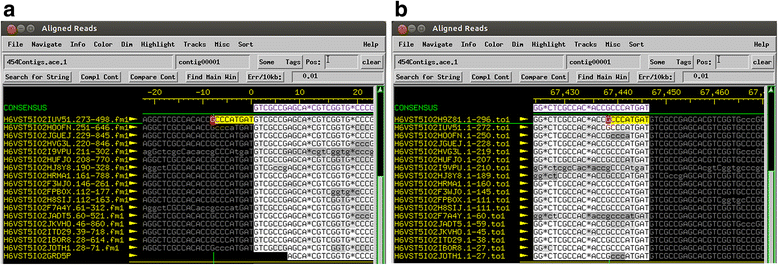
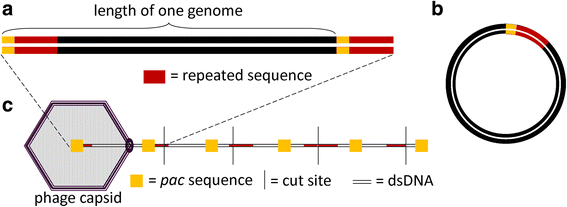
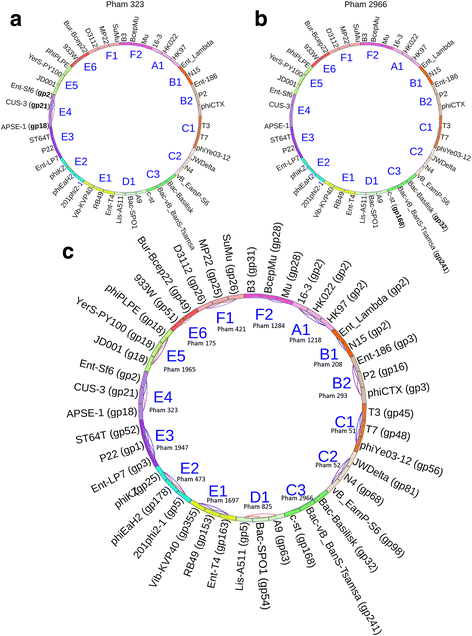
Results: The original Phamerator code can be successfully modified and custom databases can be generated using the instructions we provide. Results of genome map comparisons within a custom database reveal obstacles in performing the comparisons if a published genome has an incorrect complementarity or an incorrect location of the first base of the genome, which are common issues in GenBank-downloaded sequence files. To address these issues, we review phage packaging strategies and provide results that demonstrate identification of the genome start location and orientation using raw sequencing data and software programs such as PAUSE and Consed to establish the location of the physical ends of the genome. These results include determination of exact direct terminal repeats (DTRs) or cohesive ends, or whether phages may use a headful packaging strategy. Phylogenetic analysis using ClustalO and phamily circles in Phamerator demonstrate that the large terminase gene can be used to identify the phage packaging strategy and thereby aide in identifying the physical ends of the genome.
Conclusions: Using available online code, the Phamerator program can be customized and utilized to generate databases with individually selected genomes. These databases can then provide fruitful information in the comparative analysis of phages. Researchers can identify packaging strategies and physical ends of phage genomes using raw data from high-throughput sequencing in conjunction with phylogenetic analyses of large terminase proteins and the use of custom Phamerator databases. We promote publication of phage genomes in an orientation consistent with the physical structure of the phage chromosome and provide guidance for determining this structure.
Keywords: Comparative genomics; DNA packaging; Phage; Phamerator; Phylogenetic tree; Sequencing; Terminase.
Figures
Similar articles
-
Characterization of Paenibacillus larvae bacteriophages and their genomic relationships to firmicute bacteriophages.BMC Genomics. 2014 Aug 30;15(1):745. doi: 10.1186/1471-2164-15-745. BMC Genomics. 2014. PMID: 25174730 Free PMC article.
-
Phamerator: a bioinformatic tool for comparative bacteriophage genomics.BMC Bioinformatics. 2011 Oct 12;12:395. doi: 10.1186/1471-2105-12-395. BMC Bioinformatics. 2011. PMID: 21991981 Free PMC article.
-
Comparative genomic analysis of 142 bacteriophages infecting Salmonella enterica subsp. enterica.BMC Genomics. 2020 May 26;21(1):374. doi: 10.1186/s12864-020-6765-z. BMC Genomics. 2020. PMID: 32456612 Free PMC article.
-
A Bioinformatic Ecosystem for Bacteriophage Genomics: PhaMMSeqs, Phamerator, pdm_utils, PhagesDB, DEPhT, and PhamClust.Viruses. 2024 Aug 10;16(8):1278. doi: 10.3390/v16081278. Viruses. 2024. PMID: 39205252 Free PMC article. Review.
-
Actinobacteriophages: Genomics, Dynamics, and Applications.Annu Rev Virol. 2020 Sep 29;7(1):37-61. doi: 10.1146/annurev-virology-122019-070009. Annu Rev Virol. 2020. PMID: 32991269 Free PMC article. Review.
Cited by
-
Diversity and Ecology of Caudoviricetes Phages with Genome Terminal Repeats in Fecal Metagenomes from Four Dutch Cohorts.Viruses. 2022 Oct 20;14(10):2305. doi: 10.3390/v14102305. Viruses. 2022. PMID: 36298860 Free PMC article.
-
Characteristics of Environmental Klebsiella pneumoniae and Klebsiella oxytoca Bacteriophages and Their Therapeutic Applications.Pharmaceutics. 2023 Jan 28;15(2):434. doi: 10.3390/pharmaceutics15020434. Pharmaceutics. 2023. PMID: 36839755 Free PMC article.
-
Molecular Characterization and Taxonomic Assignment of Three Phage Isolates from a Collection Infecting Pseudomonas syringae pv. actinidiae and P. syringae pv. phaseolicola from Northern Italy.Viruses. 2021 Oct 15;13(10):2083. doi: 10.3390/v13102083. Viruses. 2021. PMID: 34696512 Free PMC article.
-
Genomic Analysis of the First European Bacteriophages with Depolymerase Activity and Biocontrol Efficacy against the Phytopathogen Ralstonia solanacearum.Viruses. 2021 Dec 17;13(12):2539. doi: 10.3390/v13122539. Viruses. 2021. PMID: 34960808 Free PMC article.
-
Isolation and in vitro characterization of novel S. epidermidis phages for therapeutic applications.Front Cell Infect Microbiol. 2023 May 24;13:1169135. doi: 10.3389/fcimb.2023.1169135. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37293203 Free PMC article.
References
-
- McAuliffe O, Ross RP, Fitzgerald GF. The new phage biology: from genomics to applications. In: McGrath S, Van Sinderen D, editors. Bacteriophage: Genetics and Molecular Biology. Norfolk, England: Caister Academic Press; 2007.
-
- Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SC, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, et al. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio. 2014;5(1):e01051–01013. doi: 10.1128/mBio.01051-13. - DOI - PMC - PubMed
-
- Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, SEA-PHAGES Program A. Modlin RL, Hendrix RW, Hatfull GF. On the nature of mycobacteriophage diversity and host preference. Virology. 2012;434(2):187–201. doi: 10.1016/j.virol.2012.09.026. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

