Structural basis for oxygen degradation domain selectivity of the HIF prolyl hydroxylases
- PMID: 27561929
- PMCID: PMC5007464
- DOI: 10.1038/ncomms12673
Structural basis for oxygen degradation domain selectivity of the HIF prolyl hydroxylases
Abstract
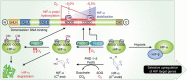
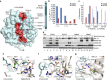
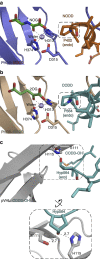
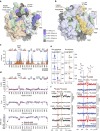
The response to hypoxia in animals involves the expression of multiple genes regulated by the αβ-hypoxia-inducible transcription factors (HIFs). The hypoxia-sensing mechanism involves oxygen limited hydroxylation of prolyl residues in the N- and C-terminal oxygen-dependent degradation domains (NODD and CODD) of HIFα isoforms, as catalysed by prolyl hydroxylases (PHD 1-3). Prolyl hydroxylation promotes binding of HIFα to the von Hippel-Lindau protein (VHL)-elongin B/C complex, thus signalling for proteosomal degradation of HIFα. We reveal that certain PHD2 variants linked to familial erythrocytosis and cancer are highly selective for CODD or NODD. Crystalline and solution state studies coupled to kinetic and cellular analyses reveal how wild-type and variant PHDs achieve ODD selectivity via different dynamic interactions involving loop and C-terminal regions. The results inform on how HIF target gene selectivity is achieved and will be of use in developing selective PHD inhibitors.
Figures


References
-
- Bruick R. K. Oxygen sensing in the hypoxic response pathway: regulation of the hypoxia-inducible transcription factor. Genes Dev. 17, 2614–2623 (2003). - PubMed
-
- Kaelin W. G. Jr & Ratcliffe P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 (2008). - PubMed
-
- Schofield C. J. & Ratcliffe P. J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell. Biol. 5, 343–354 (2004). - PubMed
-
- Semenza G. L. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 19, 176–182 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

