Metadynamic metainference: Enhanced sampling of the metainference ensemble using metadynamics
- PMID: 27561930
- PMCID: PMC4999896
- DOI: 10.1038/srep31232
Metadynamic metainference: Enhanced sampling of the metainference ensemble using metadynamics
Abstract
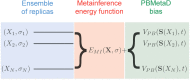
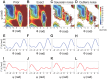
Accurate and precise structural ensembles of proteins and macromolecular complexes can be obtained with metainference, a recently proposed Bayesian inference method that integrates experimental information with prior knowledge and deals with all sources of errors in the data as well as with sample heterogeneity. The study of complex macromolecular systems, however, requires an extensive conformational sampling, which represents a separate challenge. To address such challenge and to exhaustively and efficiently generate structural ensembles we combine metainference with metadynamics and illustrate its application to the calculation of the free energy landscape of the alanine dipeptide.
Figures
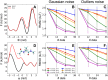
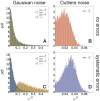
 ). When systematic errors are present, we selected two outliers (
). When systematic errors are present, we selected two outliers ( and
and  ) and two data points not affected by error (
) and two data points not affected by error ( and
and  ). When using the outliers model, we plot the distribution of the typical dataset uncertainty
). When using the outliers model, we plot the distribution of the typical dataset uncertainty  .
.
Similar articles
-
A Practical Guide to the Simultaneous Determination of Protein Structure and Dynamics Using Metainference.Methods Mol Biol. 2019;2022:313-340. doi: 10.1007/978-1-4939-9608-7_13. Methods Mol Biol. 2019. PMID: 31396909
-
Metadynamic metainference: Convergence towards force field independent structural ensembles of a disordered peptide.J Chem Phys. 2017 Apr 28;146(16):165102. doi: 10.1063/1.4981211. J Chem Phys. 2017. PMID: 28456189
-
Guide for determination of protein structural ensembles by combining cryo-EM data with metadynamics.FEBS Open Bio. 2023 Jul;13(7):1193-1203. doi: 10.1002/2211-5463.13542. Epub 2023 Jan 9. FEBS Open Bio. 2023. PMID: 36562694 Free PMC article.
-
Using simulation to interpret experimental data in terms of protein conformational ensembles.Curr Opin Struct Biol. 2017 Apr;43:79-87. doi: 10.1016/j.sbi.2016.11.018. Epub 2016 Dec 9. Curr Opin Struct Biol. 2017. PMID: 27940377 Review.
-
Advances in uncovering the mechanisms of macromolecular conformational entropy.Nat Chem Biol. 2025 May;21(5):623-634. doi: 10.1038/s41589-025-01879-3. Epub 2025 Apr 24. Nat Chem Biol. 2025. PMID: 40275100 Free PMC article. Review.
Cited by
-
Determination of the Structure and Dynamics of the Fuzzy Coat of an Amyloid Fibril of IAPP Using Cryo-Electron Microscopy.Biochemistry. 2023 Aug 15;62(16):2407-2416. doi: 10.1021/acs.biochem.3c00010. Epub 2023 Jul 21. Biochemistry. 2023. PMID: 37477459 Free PMC article.
-
A conformational fingerprint for amyloidogenic light chains.Elife. 2025 Mar 3;13:RP102002. doi: 10.7554/eLife.102002. Elife. 2025. PMID: 40028903 Free PMC article.
-
Refinement of α-Synuclein Ensembles Against SAXS Data: Comparison of Force Fields and Methods.Front Mol Biosci. 2021 Apr 22;8:654333. doi: 10.3389/fmolb.2021.654333. eCollection 2021. Front Mol Biosci. 2021. PMID: 33968988 Free PMC article.
-
Reweighting of molecular simulations with explicit-solvent SAXS restraints elucidates ion-dependent RNA ensembles.Nucleic Acids Res. 2021 Aug 20;49(14):e84. doi: 10.1093/nar/gkab459. Nucleic Acids Res. 2021. PMID: 34107023 Free PMC article.
-
Methods of probing the interactions between small molecules and disordered proteins.Cell Mol Life Sci. 2017 Sep;74(17):3225-3243. doi: 10.1007/s00018-017-2563-4. Epub 2017 Jun 19. Cell Mol Life Sci. 2017. PMID: 28631009 Free PMC article. Review.
References
-
- Rieping W., Habeck M. & Nilges M. Inferential structure determination. Science 309, 303–306 (2005). - PubMed
-
- Cavalli A., Camilloni C. & Vendruscolo M. Molecular dynamics simulations with replica-averaged structural restraints generate structural ensembles according to the maximum entropy principle. J. Chem. Phys. 138, 094112 (2013). - PubMed
-
- Lindorff-Larsen K., Best R. B., Depristo M. A., Dobson C. M. & Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature 433, 128–132 (2005). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

