Large scale free energy calculations for blind predictions of protein-ligand binding: the D3R Grand Challenge 2015
- PMID: 27562018
- PMCID: PMC5869689
- DOI: 10.1007/s10822-016-9952-x
Large scale free energy calculations for blind predictions of protein-ligand binding: the D3R Grand Challenge 2015
Abstract
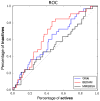
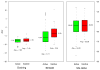
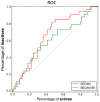
We describe binding free energy calculations in the D3R Grand Challenge 2015 for blind prediction of the binding affinities of 180 ligands to Hsp90. The present D3R challenge was built around experimental datasets involving Heat shock protein (Hsp) 90, an ATP-dependent molecular chaperone which is an important anticancer drug target. The Hsp90 ATP binding site is known to be a challenging target for accurate calculations of ligand binding affinities because of the ligand-dependent conformational changes in the binding site, the presence of ordered waters and the broad chemical diversity of ligands that can bind at this site. Our primary focus here is to distinguish binders from nonbinders. Large scale absolute binding free energy calculations that cover over 3000 protein-ligand complexes were performed using the BEDAM method starting from docked structures generated by Glide docking. Although the ligand dataset in this study resembles an intermediate to late stage lead optimization project while the BEDAM method is mainly developed for early stage virtual screening of hit molecules, the BEDAM binding free energy scoring has resulted in a moderate enrichment of ligand screening against this challenging drug target. Results show that, using a statistical mechanics based free energy method like BEDAM starting from docked poses offers better enrichment than classical docking scoring functions and rescoring methods like Prime MM-GBSA for the Hsp90 data set in this blind challenge. Importantly, among the three methods tested here, only the mean value of the BEDAM binding free energy scores is able to separate the large group of binders from the small group of nonbinders with a gap of 2.4 kcal/mol. None of the three methods that we have tested provided accurate ranking of the affinities of the 147 active compounds. We discuss the possible sources of errors in the binding free energy calculations. The study suggests that BEDAM can be used strategically to discriminate binders from nonbinders in virtual screening and to more accurately predict the ligand binding modes prior to the more computationally expensive FEP calculations of binding affinity.
Keywords: Binding free energy; D3R; Docking; GC2015; Hsp90; ROC.
Figures

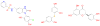
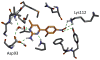

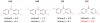
Similar articles
-
Binding-affinity predictions of HSP90 in the D3R Grand Challenge 2015 with docking, MM/GBSA, QM/MM, and free-energy simulations.J Comput Aided Mol Des. 2016 Sep;30(9):707-730. doi: 10.1007/s10822-016-9942-z. Epub 2016 Aug 26. J Comput Aided Mol Des. 2016. PMID: 27565797 Free PMC article.
-
Blinded predictions of binding modes and energies of HSP90-α ligands for the 2015 D3R grand challenge.Bioorg Med Chem. 2016 Oct 15;24(20):4890-4899. doi: 10.1016/j.bmc.2016.07.044. Epub 2016 Jul 21. Bioorg Med Chem. 2016. PMID: 27485604
-
Predicting binding poses and affinities for protein - ligand complexes in the 2015 D3R Grand Challenge using a physical model with a statistical parameter estimation.J Comput Aided Mol Des. 2016 Sep;30(9):791-804. doi: 10.1007/s10822-016-9976-2. Epub 2016 Oct 7. J Comput Aided Mol Des. 2016. PMID: 27718029
-
Advances in Docking.Curr Med Chem. 2019;26(42):7555-7580. doi: 10.2174/0929867325666180904115000. Curr Med Chem. 2019. PMID: 30182836 Review.
-
Free energy calculations to estimate ligand-binding affinities in structure-based drug design.Curr Pharm Des. 2014;20(20):3323-37. doi: 10.2174/13816128113199990604. Curr Pharm Des. 2014. PMID: 23947646 Review.
Cited by
-
Docking of small molecules to farnesoid X receptors using AutoDock Vina with the Convex-PL potential: lessons learned from D3R Grand Challenge 2.J Comput Aided Mol Des. 2018 Jan;32(1):151-162. doi: 10.1007/s10822-017-0062-1. Epub 2017 Sep 14. J Comput Aided Mol Des. 2018. PMID: 28913782
-
Optimal affinity ranking for automated virtual screening validated in prospective D3R grand challenges.J Comput Aided Mol Des. 2018 Jan;32(1):287-297. doi: 10.1007/s10822-017-0065-y. Epub 2017 Sep 16. J Comput Aided Mol Des. 2018. PMID: 28918599 Free PMC article.
-
Improving small molecule virtual screening strategies for the next generation of therapeutics.Curr Opin Chem Biol. 2018 Jun;44:87-92. doi: 10.1016/j.cbpa.2018.06.006. Epub 2018 Jun 17. Curr Opin Chem Biol. 2018. PMID: 29920436 Free PMC article. Review.
-
Resolving the Ligand-Binding Specificity in c-MYC G-Quadruplex DNA: Absolute Binding Free Energy Calculations and SPR Experiment.J Phys Chem B. 2017 Nov 22;121(46):10484-10497. doi: 10.1021/acs.jpcb.7b09406. Epub 2017 Nov 9. J Phys Chem B. 2017. PMID: 29086571 Free PMC article.
-
Computation of Protein-Ligand Binding Free Energies with a Quantum Mechanics-Based Mining Minima Algorithm.J Chem Theory Comput. 2025 Apr 22;21(8):4236-4265. doi: 10.1021/acs.jctc.4c01707. Epub 2025 Apr 9. J Chem Theory Comput. 2025. PMID: 40202178 Free PMC article.
References
-
- Boresch S, Tettinger F, Leitgeb M, Karplus M. Absolute binding free energies: a quantitative approach for their calculation. J Phys Chem. 2003;107:9535–9551.
-
- Chang C-E, Gilson MK. Free energy, entropy, and induced fit in host-guest recognition: calculations with the second-generation mining minima algorithm. J Am Chem Soc. 2004;126:13156–13164. - PubMed
-
- Deng Y, Roux B. Calculation of standard binding free energies: aromatic molecules in the T4 lysozyme L99A mutant. J Chem Theory Comput. 2006;2:1255–1273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

