Na+ coordination at the Na2 site of the Na+/I- symporter
- PMID: 27562170
- PMCID: PMC5027462
- DOI: 10.1073/pnas.1607231113
Na+ coordination at the Na2 site of the Na+/I- symporter
Abstract
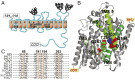
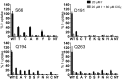
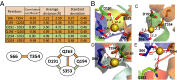
The sodium/iodide symporter (NIS) mediates active I(-) transport in the thyroid-the first step in thyroid hormone biosynthesis-with a 2 Na(+): 1 I(-) stoichiometry. The two Na(+) binding sites (Na1 and Na2) and the I(-) binding site interact allosterically: when Na(+) binds to a Na(+) site, the affinity of NIS for the other Na(+) and for I(-) increases significantly. In all Na(+)-dependent transporters with the same fold as NIS, the side chains of two residues, S353 and T354 (NIS numbering), were identified as the Na(+) ligands at Na2. To understand the cooperativity between the substrates, we investigated the coordination at the Na2 site. We determined that four other residues-S66, D191, Q194, and Q263-are also involved in Na(+) coordination at this site. Experiments in whole cells demonstrated that these four residues participate in transport by NIS: mutations at these positions result in proteins that, although expressed at the plasma membrane, transport little or no I(-) These residues are conserved throughout the entire SLC5 family, to which NIS belongs, suggesting that they serve a similar function in the other transporters. Our findings also suggest that the increase in affinity that each site displays when an ion binds to another site may result from changes in the dynamics of the transporter. These mechanistic insights deepen our understanding not only of NIS but also of other transporters, including many that, like NIS, are of great medical relevance.
Keywords: NIS; Na+ binding site; Na+-driven cotransporters; Na+/I− symporter; protein dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
The Iodide Transport Defect-Causing Y348D Mutation in the Na+/I- Symporter Renders the Protein Intrinsically Inactive and Impairs Its Targeting to the Plasma Membrane.Thyroid. 2021 Aug;31(8):1272-1281. doi: 10.1089/thy.2020.0931. Epub 2021 Jun 4. Thyroid. 2021. PMID: 33779310 Free PMC article.
-
The sodium/iodide symporter: state of the art of its molecular characterization.Biochim Biophys Acta. 2014 Jan;1838(1 Pt B):244-53. doi: 10.1016/j.bbamem.2013.08.013. Epub 2013 Aug 27. Biochim Biophys Acta. 2014. PMID: 23988430 Review.
-
The Na+/I- symporter (NIS): recent advances.J Bioenerg Biomembr. 1998 Apr;30(2):195-206. doi: 10.1023/a:1020577426732. J Bioenerg Biomembr. 1998. PMID: 9672241 Review.
-
Amino acid residues in transmembrane segment IX of the Na+/I- symporter play a role in its Na+ dependence and are critical for transport activity.J Biol Chem. 2007 Aug 31;282(35):25290-8. doi: 10.1074/jbc.M700147200. Epub 2007 Jul 2. J Biol Chem. 2007. PMID: 17606623
-
Beyond non-integer Hill coefficients: A novel approach to analyzing binding data, applied to Na+-driven transporters.J Gen Physiol. 2015 Jun;145(6):555-63. doi: 10.1085/jgp.201511365. J Gen Physiol. 2015. PMID: 26009546 Free PMC article.
Cited by
-
Allosteric regulation of mammalian Na+/I- symporter activity by perchlorate.Nat Struct Mol Biol. 2020 Jun;27(6):533-539. doi: 10.1038/s41594-020-0417-5. Epub 2020 May 25. Nat Struct Mol Biol. 2020. PMID: 32451489 Free PMC article.
-
Iodide Binding in Sodium-Coupled Cotransporters.J Chem Inf Model. 2017 Dec 26;57(12):3043-3055. doi: 10.1021/acs.jcim.7b00521. Epub 2017 Dec 4. J Chem Inf Model. 2017. PMID: 29131623 Free PMC article.
-
A Novel SLC5A5 Variant Reveals the Crucial Role of Kinesin Light Chain 2 in Thyroid Hormonogenesis.J Clin Endocrinol Metab. 2021 Jun 16;106(7):1867-1881. doi: 10.1210/clinem/dgab283. J Clin Endocrinol Metab. 2021. PMID: 33912899 Free PMC article.
-
Targeted Next-Generation Sequencing of Congenital Hypothyroidism-Causative Genes Reveals Unexpected Thyroglobulin Gene Variants in Patients with Iodide Transport Defect.Int J Mol Sci. 2022 Aug 17;23(16):9251. doi: 10.3390/ijms23169251. Int J Mol Sci. 2022. PMID: 36012511 Free PMC article.
-
Effects of a Phosphoinositide-3-Kinase Inhibitor on Anaplastic Thyroid Cancer Stem Cells.Asian Pac J Cancer Prev. 2017 Aug 27;18(8):2287-2291. doi: 10.22034/APJCP.2017.18.8.2287. Asian Pac J Cancer Prev. 2017. PMID: 28843268 Free PMC article.
References
-
- Carrasco N. Iodide transport in the thyroid gland. Biochim Biophys Acta. 1993;1154(1):65–82. - PubMed
-
- Dai G, Levy O, Carrasco N. Cloning and characterization of the thyroid iodide transporter. Nature. 1996;379(6564):458–460. - PubMed
-
- De La Vieja A, Dohan O, Levy O, Carrasco N. Molecular analysis of the sodium/iodide symporter: Impact on thyroid and extrathyroid pathophysiology. Physiol Rev. 2000;80(3):1083–1105. - PubMed
-
- Eskandari S, et al. Thyroid Na+/I− symporter. Mechanism, stoichiometry, and specificity. J Biol Chem. 1997;272(43):27230–27238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

