Structures of Ebola virus GP and sGP in complex with therapeutic antibodies
- PMID: 27562261
- PMCID: PMC5003320
- DOI: 10.1038/nmicrobiol.2016.128
Structures of Ebola virus GP and sGP in complex with therapeutic antibodies
Abstract
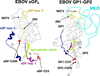
The Ebola virus (EBOV) GP gene encodes two glycoproteins. The major product is a soluble, dimeric glycoprotein (sGP) that is secreted abundantly. Despite the abundance of sGP during infection, little is known regarding its structure or functional role. A minor product, resulting from transcriptional editing, is the transmembrane-anchored, trimeric viral surface glycoprotein (GP). GP mediates attachment to and entry into host cells, and is the intended target of antibody therapeutics. Because large portions of sequence are shared between GP and sGP, it has been hypothesized that sGP may potentially subvert the immune response or may contribute to pathogenicity. In this study, we present cryo-electron microscopy structures of GP and sGP in complex with GP-specific and GP/sGP cross-reactive antibodies undergoing human clinical trials. The structure of the sGP dimer presented here, in complex with both an sGP-specific antibody and a GP/sGP cross-reactive antibody, permits us to unambiguously assign the oligomeric arrangement of sGP and compare its structure and epitope presentation to those of GP. We also provide biophysical evaluation of naturally occurring GP/sGP mutations that fall within the footprints identified by our high-resolution structures. Taken together, our data provide a detailed and more complete picture of the accessible Ebolavirus glycoprotein landscape and a structural basis to evaluate patient and vaccine antibody responses towards differently structured products of the GP gene.
Figures
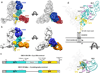



Similar articles
-
Characterization of Immune Responses Induced by Ebola Virus Glycoprotein (GP) and Truncated GP Isoform DNA Vaccines and Protection Against Lethal Ebola Virus Challenge in Mice.J Infect Dis. 2015 Oct 1;212 Suppl 2(Suppl 2):S398-403. doi: 10.1093/infdis/jiv186. Epub 2015 Apr 14. J Infect Dis. 2015. PMID: 25877553 Free PMC article.
-
The Roles of Ebola Virus Soluble Glycoprotein in Replication, Pathogenesis, and Countermeasure Development.Viruses. 2019 Oct 31;11(11):999. doi: 10.3390/v11110999. Viruses. 2019. PMID: 31683550 Free PMC article. Review.
-
Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus.PLoS Pathog. 2012;8(12):e1003065. doi: 10.1371/journal.ppat.1003065. Epub 2012 Dec 13. PLoS Pathog. 2012. PMID: 23271969 Free PMC article.
-
RNA Editing of the GP Gene of Ebola Virus is an Important Pathogenicity Factor.J Infect Dis. 2015 Oct 1;212 Suppl 2:S226-33. doi: 10.1093/infdis/jiv309. Epub 2015 Jul 2. J Infect Dis. 2015. PMID: 26138826
-
[Research progress on ebola virus glycoprotein].Bing Du Xue Bao. 2013 Mar;29(2):233-7. Bing Du Xue Bao. 2013. PMID: 23757858 Review. Chinese.
Cited by
-
Structural basis for broad neutralization of ebolaviruses by an antibody targeting the glycoprotein fusion loop.Nat Commun. 2018 Sep 26;9(1):3934. doi: 10.1038/s41467-018-06113-4. Nat Commun. 2018. PMID: 30258051 Free PMC article.
-
Fighting Ebola: A Window for Vaccine Re-evaluation?PLoS Pathog. 2017 Jan 12;13(1):e1006037. doi: 10.1371/journal.ppat.1006037. eCollection 2017 Jan. PLoS Pathog. 2017. PMID: 28081225 Free PMC article. Review. No abstract available.
-
Antibody-mediated protection against Ebola virus.Nat Immunol. 2018 Nov;19(11):1169-1178. doi: 10.1038/s41590-018-0233-9. Epub 2018 Oct 17. Nat Immunol. 2018. PMID: 30333617 Free PMC article. Review.
-
Thermostable Ebola virus vaccine formulations lyophilized in the presence of aluminum hydroxide.Eur J Pharm Biopharm. 2019 Mar;136:213-220. doi: 10.1016/j.ejpb.2019.01.019. Epub 2019 Jan 28. Eur J Pharm Biopharm. 2019. PMID: 30703544 Free PMC article.
-
Extracellular Vesicles and Ebola Virus: A New Mechanism of Immune Evasion.Viruses. 2019 May 2;11(5):410. doi: 10.3390/v11050410. Viruses. 2019. PMID: 31052499 Free PMC article. Review.
References
-
- Henao-Restrepo AM, et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet. 2015;386:857–866. - PubMed
-
- Stanley DA, et al. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nature medicine. 2014;20:1126–1129. - PubMed
-
- Tapia MD, et al. Use of ChAd3-EBO-Z Ebola virus vaccine in Malian and US adults, and boosting of Malian adults with MVA-BN-Filo: a phase 1, single-blind, randomised trial, a phase 1b, open-label and double-blind, dose-escalation trial, and a nested, randomised, double-blind, placebo-controlled trial. Lancet Infect Dis. 2016;16:31–42. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

