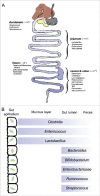
Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model
- PMID: 27562711
- PMCID: PMC5411234
- DOI: 10.1080/21505594.2016.1208890
Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model
Abstract
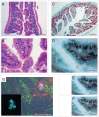
The mammalian gastrointestinal (GI) tract is a complex organ system with a twist-a significant portion of its composition is a community of microbial symbionts. The microbiota plays an increasingly appreciated role in many clinically-relevant conditions. It is important to understand the details of biofilm development in the GI tract since bacteria in this state not only use biofilms to improve colonization, biofilm bacteria often exhibit high levels of resistance to common, clinically relevant antibacterial drugs. Here we examine the initial colonization of the germ-free murine GI tract by Enterococcus faecalis-one of the first bacterial colonizers of the naïve mammalian gut. We demonstrate strong morphological similarities to our previous in vitro E. faecalis biofilm microcolony architecture using 3 complementary imaging techniques: conventional tissue Gram stain, immunofluorescent imaging (IFM) of constitutive fluorescent protein reporter expression, and low-voltage scanning electron microscopy (LV-SEM). E. faecalis biofilm microcolonies were readily identifiable throughout the entire lower GI tract, from the duodenum to the colon. Notably, biofilm development appeared to occur as discrete microcolonies directly attached to the epithelial surface rather than confluent sheets of cells throughout the GI tract even in the presence of high (>109) fecal bacterial loads. An in vivo competition experiment using a pool of 11 select E. faecalis mutant strains containing sequence-defined transposon insertions showed the potential of this model to identify genetic factors involved in E. faecalis colonization of the murine GI tract.
Keywords: antibiotic resistance; competitive fitness; intestinal microbiota; opportunistic pathogen.
Figures



References
-
- Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev [Internet] 2010. [cited 2014December17]; 90:859-904. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20664075; PMID:20664075; http://dx.doi.org/ 10.1152/physrev.00045.2009 - DOI - PubMed
-
- Quigley EMM. Small intestinal bacterial overgrowth: what it is and what it is not. Curr Opin Gastroenterol [Internet] 2014. [cited 2016January4]; 30:141-6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24406476; PMID:24406476; http://dx.doi.org/ 10.1097/MOG.0000000000000040 - DOI - PubMed
-
- Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol [Internet] 1996. [cited 2016January7]; 4:430-5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8950812; PMID:8950812; http://dx.doi.org/ 10.1016/0966-842X(96)10057-3 - DOI - PubMed
-
- Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med [Internet] 1968. [cited 2015December31]; 127:67-76. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2138434&tool=p...; PMID:4169441; http://dx.doi.org/ 10.1084/jem.127.1.67 - DOI - PMC - PubMed
-
- Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol [Internet] 2013. [cited 2014September15]; 14:660-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23778793; PMID:23778793; http://dx.doi.org/ 10.1038/ni.2611 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
