3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM
- PMID: 27563056
- PMCID: PMC5041491
- DOI: 10.1093/nar/gkw708
3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM
Abstract
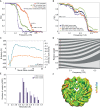
The Volta phase plate is a recently developed electron cryo-microscopy (cryo-EM) device that enables contrast enhancement of biological samples. Here we have evaluated the potential of combining phase-plate imaging and single particle analysis to determine the structure of a small protein-DNA complex. To test the method, we made use of a 200 kDa Nucleosome Core Particle (NCP) reconstituted with 601 DNA for which a high-resolution X-ray crystal structure is known. We find that the phase plate provides a significant contrast enhancement that permits individual NCPs and DNA to be clearly identified in amorphous ice. The refined structure from 26,060 particles has an overall resolution of 3.9 Å and the density map exhibits structural features consistent with the estimated resolution, including clear density for amino acid side chains and DNA features such as the phosphate backbone. Our results demonstrate that phase-plate cryo-EM promises to become an important method to determine novel near-atomic resolution structures of small and challenging samples, such as nucleosomes in complex with nucleosome-binding factors.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Bai X.C., McMullan G., Scheres S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. - PubMed
-
- Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995;28:171–193. - PubMed
-
- Baker L.A., Rubinstein J.L. Radiation damage in electron cryomicroscopy. Methods Enzymol. 2010;481:371–388. - PubMed

