Targeted Disruption of the Interaction between WD-40 Repeat Protein 5 (WDR5) and Mixed Lineage Leukemia (MLL)/SET1 Family Proteins Specifically Inhibits MLL1 and SETd1A Methyltransferase Complexes
- PMID: 27563068
- PMCID: PMC5077178
- DOI: 10.1074/jbc.M116.752626
Targeted Disruption of the Interaction between WD-40 Repeat Protein 5 (WDR5) and Mixed Lineage Leukemia (MLL)/SET1 Family Proteins Specifically Inhibits MLL1 and SETd1A Methyltransferase Complexes
Abstract
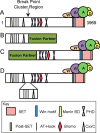
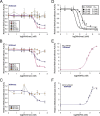
MLL1 belongs to the SET1 family of histone H3 lysine 4 (H3K4) methyltransferases, composed of MLL1-4 and SETd1A/B. MLL1 translocations are present in acute leukemias, and mutations in several family members are associated with cancer and developmental disorders. MLL1 associates with a subcomplex containing WDR5, RbBP5, ASH2L, and DPY-30 (WRAD), forming the MLL1 core complex required for H3K4 mono- and dimethylation and transcriptional activation. Core complex assembly requires interaction of WDR5 with the MLL1 Win (WDR5 interaction) motif, which is conserved across the SET1 family. Agents that mimic the SET1 family Win motif inhibit the MLL1 core complex and have become an attractive approach for targeting MLL1 in cancers. Like MLL1, other SET1 family members interact with WRAD, but the roles of the Win motif in complex assembly and enzymatic activity remain unexplored. Here, we show that the Win motif is necessary for interaction of WDR5 with all members of the human SET1 family. Mutation of the Win motif-WDR5 interface severely disrupts assembly and activity of MLL1 and SETd1A complexes but only modestly disrupts MLL2/4 and SETd1B complexes without significantly altering enzymatic activity in vitro Notably, in the absence of WDR5, MLL3 interacts with RAD and shows enhanced activity. To further probe the role of the Win motif-WDR5 interaction, we designed a peptidomimetic that binds WDR5 (Kd ∼3 nm) and selectively inhibits activity of MLL1 and SETd1A core complexes within the SET1 family. Our results reveal that SET1 family complexes with the weakest Win motif-WDR5 interaction are more susceptible to Win motif-based inhibitors.
Keywords: MLL1; SETd1A; WDR5; Win motif; enzyme; epigenetics; histone methylation; protein structure; protein-protein interaction.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Lee J. H., Tate C. M., You J. S., and Skalnik D. G. (2007) Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 282, 13419–13428 - PubMed
-
- Lee J. H., and Skalnik D. G. (2005) CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 280, 41725–41731 - PubMed
-
- Ruault M., Brun M. E., Ventura M., Roizès G., and De Sario A. (2002) MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene 284, 73–81 - PubMed
-
- Huntsman D. G., Chin S. F., Muleris M., Batley S. J., Collins V. P., Wiedemann L. M., Aparicio S., and Caldas C. (1999) MLL2, the second human homolog of the Drosophila trithorax gene, maps to 19q13.1 and is amplified in solid tumor cell lines. Oncogene 18, 7975–7984 - PubMed
-
- Prasad R., Zhadanov A. B., Sedkov Y., Bullrich F., Druck T., Rallapalli R., Yano T., Alder H., Croce C. M., Huebner K., Mazo A., and Canaani E. (1997) Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene 15, 549–560 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

