Epigenetic Control of Defense Signaling and Priming in Plants
- PMID: 27563304
- PMCID: PMC4980392
- DOI: 10.3389/fpls.2016.01201
Epigenetic Control of Defense Signaling and Priming in Plants
Abstract
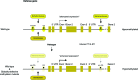
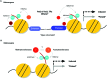
Immune recognition of pathogen-associated molecular patterns or effectors leads to defense activation at the pathogen challenged sites. This is followed by systemic defense activation at distant non-challenged sites, termed systemic acquired resistance (SAR). These inducible defenses are accompanied by extensive transcriptional reprogramming of defense-related genes. SAR is associated with priming, in which a subset of these genes is kept at a poised state to facilitate subsequent transcriptional regulation. Transgenerational inheritance of defense-related priming in plants indicates the stability of such primed states. Recent studies have revealed the importance and dynamic engagement of epigenetic mechanisms, such as DNA methylation and histone modifications that are closely linked to chromatin reconfiguration, in plant adaptation to different biotic stresses. Herein we review current knowledge regarding the biological significance and underlying mechanisms of epigenetic control for immune responses in plants. We also argue for the importance of host transposable elements as critical regulators of interactions in the evolutionary "arms race" between plants and pathogens.
Keywords: DNA methylation; defense priming; epigenetic control; histone modification; plant immunity; plant-microbe interactions; transposable elements.
Figures
Similar articles
-
Epigenetic Mechanisms: An Emerging Player in Plant-Microbe Interactions.Mol Plant Microbe Interact. 2016 Mar;29(3):187-96. doi: 10.1094/MPMI-08-15-0194-FI. Epub 2016 Feb 5. Mol Plant Microbe Interact. 2016. PMID: 26524162 Review.
-
Plant Immunity: From Signaling to Epigenetic Control of Defense.Trends Plant Sci. 2018 Sep;23(9):833-844. doi: 10.1016/j.tplants.2018.06.004. Epub 2018 Jun 30. Trends Plant Sci. 2018. PMID: 29970339 Review.
-
Epigenetic weapons of plants against fungal pathogens.BMC Plant Biol. 2024 Mar 6;24(1):175. doi: 10.1186/s12870-024-04829-8. BMC Plant Biol. 2024. PMID: 38443788 Free PMC article. Review.
-
Priming of antiherbivore defensive responses in plants.Insect Sci. 2013 Jun;20(3):273-85. doi: 10.1111/j.1744-7917.2012.01584.x. Epub 2012 Dec 19. Insect Sci. 2013. PMID: 23955880 Review.
-
Priming for enhanced defense.Annu Rev Phytopathol. 2015;53:97-119. doi: 10.1146/annurev-phyto-080614-120132. Epub 2015 Jun 11. Annu Rev Phytopathol. 2015. PMID: 26070330 Review.
Cited by
-
Can Forest Trees Cope with Climate Change?-Effects of DNA Methylation on Gene Expression and Adaptation to Environmental Change.Int J Mol Sci. 2021 Dec 16;22(24):13524. doi: 10.3390/ijms222413524. Int J Mol Sci. 2021. PMID: 34948318 Free PMC article. Review.
-
Epigenetic Patterns and Geographical Parthenogenesis in the Alpine Plant Species Ranunculus kuepferi (Ranunculaceae).Int J Mol Sci. 2020 May 7;21(9):3318. doi: 10.3390/ijms21093318. Int J Mol Sci. 2020. PMID: 32392879 Free PMC article.
-
Sequence Composition of Bacterial Chromosome Clones in a Transgressive Root-Knot Nematode Resistance Chromosome Region in Tetraploid Cotton.Front Plant Sci. 2020 Dec 14;11:574486. doi: 10.3389/fpls.2020.574486. eCollection 2020. Front Plant Sci. 2020. PMID: 33381129 Free PMC article.
-
POWERDRESS positively regulates systemic acquired resistance in Arabidopsis.Plant Cell Rep. 2022 Dec;41(12):2351-2362. doi: 10.1007/s00299-022-02926-2. Epub 2022 Sep 24. Plant Cell Rep. 2022. PMID: 36152035
-
Transcriptomic and methylation analysis of susceptible and tolerant grapevine genotypes following Plasmopara viticola infection.Physiol Plant. 2022 Sep;174(5):e13771. doi: 10.1111/ppl.13771. Physiol Plant. 2022. PMID: 36053855 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

