Mitochondrial-Targeting MET Kinase Inhibitor Kills Erlotinib-Resistant Lung Cancer Cells
- PMID: 27563407
- PMCID: PMC4983733
- DOI: 10.1021/acsmedchemlett.6b00223
Mitochondrial-Targeting MET Kinase Inhibitor Kills Erlotinib-Resistant Lung Cancer Cells
Abstract
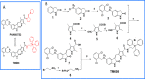
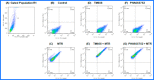
Lung cancer cells harboring activating EGFR mutations acquire resistance to EGFR tyrosine kinase inhibitors (TKIs) by activating several bypass mechanisms, including MET amplification and overexpression. We show that a significant proportion of activated MET protein in EGFR TKI-resistant HCC827 lung cancer cells resides within the mitochondria. Targeting the total complement of MET in the plasma membrane and mitochondria should render these cells more susceptible to cell death and hence provide a means of circumventing drug resistance. Herein, the mitochondrial targeting triphenylphosphonium (TPP) moiety was introduced to the selective MET kinase inhibitor PHA665752. The resulting TPP analogue rapidly localized to the mitochondria of MET-overexpressing erlotinib-resistant HCC827 cells, partially suppressed the phosphorylation (Y1234/Y1235) of MET in the mitochondrial inner membrane and was as cytotoxic and apoptogenic as the parent compound. These findings provide support for the targeting of mitochondrial MET with a TPP-TKI conjugate as a means of restoring responsiveness to chemotherapy.
Keywords: MET kinase; Mitochondrial targeting; PHA665752; drug resistance; nonsmall cell lung cancer; triphenylphosphonium conjugate.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Wilson T. R.; Fridlyand J.; Yan Y.; Penuel E.; Burton L.; Chan E.; Peng J.; Lin E.; Wang Y.; Sosman J.; Ribas A.; Li J.; Moffat J.; Sutherlin D. P.; Koeppen H.; Merchant M.; Neve R.; Settleman J. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012, 487, 505–509. 10.1038/nature11249. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

