Smc5/6 Is a Telomere-Associated Complex that Regulates Sir4 Binding and TPE
- PMID: 27564449
- PMCID: PMC5001636
- DOI: 10.1371/journal.pgen.1006268
Smc5/6 Is a Telomere-Associated Complex that Regulates Sir4 Binding and TPE
Abstract
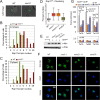
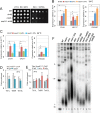
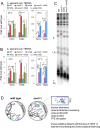
SMC proteins constitute the core members of the Smc5/6, cohesin and condensin complexes. We demonstrate that Smc5/6 is present at telomeres throughout the cell cycle and its association with chromosome ends is dependent on Nse3, a subcomponent of the complex. Cells harboring a temperature sensitive mutant, nse3-1, are defective in Smc5/6 localization to telomeres and have slightly shorter telomeres. Nse3 interacts physically and genetically with two Rap1-binding factors, Rif2 and Sir4. Reduction in telomere-associated Smc5/6 leads to defects in telomere clustering, dispersion of the silencing factor, Sir4, and a loss in transcriptional repression for sub-telomeric genes and non-coding telomeric repeat-containing RNA (TERRA). SIR4 recovery at telomeres is reduced in cells lacking Smc5/6 functionality and vice versa. However, nse3-1/ sir4 Δ double mutants show additive defects for telomere shortening and TPE indicating the contribution of Smc5/6 to telomere homeostasis is only in partial overlap with SIR factor silencing. These findings support a role for Smc5/6 in telomere maintenance that is separate from its canonical role(s) in HR-mediated events during replication and telomere elongation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Abrupt telomere losses and reduced end-resection can explain accelerated senescence of Smc5/6 mutants lacking telomerase.DNA Repair (Amst). 2011 Mar 7;10(3):271-82. doi: 10.1016/j.dnarep.2010.11.010. Epub 2010 Dec 28. DNA Repair (Amst). 2011. PMID: 21190904
-
Ku Binding on Telomeres Occurs at Sites Distal from the Physical Chromosome Ends.PLoS Genet. 2016 Dec 8;12(12):e1006479. doi: 10.1371/journal.pgen.1006479. eCollection 2016 Dec. PLoS Genet. 2016. PMID: 27930670 Free PMC article.
-
The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time.EMBO J. 2006 Apr 5;25(7):1505-14. doi: 10.1038/sj.emboj.7601038. Epub 2006 Mar 9. EMBO J. 2006. PMID: 16525505 Free PMC article.
-
Taking cohesin and condensin in context.PLoS Genet. 2018 Jan 25;14(1):e1007118. doi: 10.1371/journal.pgen.1007118. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29370184 Free PMC article. Review.
-
Functional interplay between cohesin and Smc5/6 complexes.Chromosoma. 2014 Oct;123(5):437-45. doi: 10.1007/s00412-014-0474-9. Epub 2014 Jul 1. Chromosoma. 2014. PMID: 24981336 Free PMC article. Review.
Cited by
-
SIR telomere silencing depends on nuclear envelope lipids and modulates sensitivity to a lysolipid.J Cell Biol. 2023 Jul 3;222(7):e202206061. doi: 10.1083/jcb.202206061. Epub 2023 Apr 12. J Cell Biol. 2023. PMID: 37042812 Free PMC article.
-
The Smc5/6 complex regulates the yeast Mph1 helicase at RNA-DNA hybrid-mediated DNA damage.PLoS Genet. 2017 Dec 27;13(12):e1007136. doi: 10.1371/journal.pgen.1007136. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29281624 Free PMC article.
-
Hepatitis B Viral Protein HBx: Roles in Viral Replication and Hepatocarcinogenesis.Viruses. 2024 Aug 26;16(9):1361. doi: 10.3390/v16091361. Viruses. 2024. PMID: 39339838 Free PMC article. Review.
-
TERRA expression is regulated by the telomere-binding proteins POT-1 and POT-2 in Caenorhabditis elegans.Nucleic Acids Res. 2023 Oct 27;51(19):10681-10699. doi: 10.1093/nar/gkad742. Nucleic Acids Res. 2023. PMID: 37713629 Free PMC article.
-
Live-cell imaging reveals the dynamics and function of single-telomere TERRA molecules in cancer cells.RNA Biol. 2018;15(6):787-796. doi: 10.1080/15476286.2018.1456300. Epub 2018 Apr 16. RNA Biol. 2018. PMID: 29658398 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

