Antimicrobial Peptide Resistance Genes in the Plant Pathogen Dickeya dadantii
- PMID: 27565623
- PMCID: PMC5066359
- DOI: 10.1128/AEM.01757-16
Antimicrobial Peptide Resistance Genes in the Plant Pathogen Dickeya dadantii
Abstract
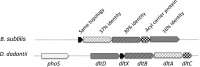
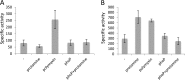
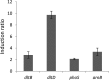
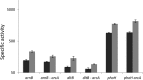
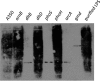
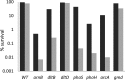
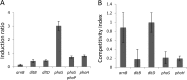
Modification of teichoic acid through the incorporation of d-alanine confers resistance in Gram-positive bacteria to antimicrobial peptides (AMPs). This process involves the products of the dltXABCD genes. These genes are widespread in Gram-positive bacteria, and they are also found in a few Gram-negative bacteria. Notably, these genes are present in all soft-rot enterobacteria (Pectobacterium and Dickeya) whose dltDXBAC operons have been sequenced. We studied the function and regulation of these genes in Dickeya dadantii dltB expression was induced in the presence of the AMP polymyxin. It was not regulated by PhoP, which controls the expression of some genes involved in AMP resistance, but was regulated by ArcA, which has been identified as an activator of genes involved in AMP resistance. However, arcA was not the regulator responsible for polymyxin induction of these genes in this bacterium, which underlines the complexity of the mechanisms controlling AMP resistance in D. dadantii Two other genes involved in resistance to AMPs have also been characterized, phoS and phoH dltB, phoS, phoH, and arcA but not dltD mutants were more sensitive to polymyxin than the wild-type strain. Decreased fitness of the dltB, phoS, and phoH mutants in chicory leaves indicates that their products are important for resistance to plant AMPs.
Importance: Gram-negative bacteria can modify their lipopolysaccharides (LPSs) to resist antimicrobial peptides (AMPs). Soft-rot enterobacteria (Dickeya and Pectobacterium spp.) possess homologues of the dlt genes in their genomes which, in Gram-positive bacteria, are involved in resistance to AMPs. In this study, we show that these genes confer resistance to AMPs, probably by modifying LPSs, and that they are required for the fitness of the bacteria during plant infection. Two other new genes involved in resistance were also analyzed. These results show that bacterial resistance to AMPs can occur in bacteria through many different mechanisms that need to be characterized.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

