Down-regulation of GIGANTEA-like genes increases plant growth and salt stress tolerance in poplar
- PMID: 27565626
- PMCID: PMC5316923
- DOI: 10.1111/pbi.12628
Down-regulation of GIGANTEA-like genes increases plant growth and salt stress tolerance in poplar
Abstract
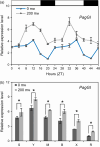
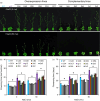
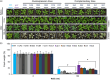
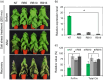
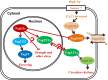
The flowering time regulator GIGANTEA (GI) connects networks involved in developmental stage transitions and environmental stress responses in Arabidopsis. However, little is known about the role of GI in growth, development and responses to environmental challenges in the perennial plant poplar. Here, we identified and functionally characterized three GI-like genes (PagGIa, PagGIb and PagGIc) from poplar (Populus alba × Populus glandulosa). PagGIs are predominantly nuclear localized and their transcripts are rhythmically expressed, with a peak around zeitgeber time 12 under long-day conditions. Overexpressing PagGIs in wild-type (WT) Arabidopsis induced early flowering and salt sensitivity, while overexpressing PagGIs in the gi-2 mutant completely or partially rescued its delayed flowering and enhanced salt tolerance phenotypes. Furthermore, the PagGIs-PagSOS2 complexes inhibited PagSOS2-regulated phosphorylation of PagSOS1 in the absence of stress, whereas these inhibitions were eliminated due to the degradation of PagGIs under salt stress. Down-regulation of PagGIs by RNA interference led to vigorous growth, higher biomass and enhanced salt stress tolerance in transgenic poplar plants. Taken together, these results indicate that several functions of Arabidopsis GI are conserved in its poplar orthologues, and they lay the foundation for developing new approaches to producing salt-tolerant trees for sustainable development on marginal lands worldwide.
Keywords: Arabidopsis; PagGI; RNAi; poplar; salt tolerance; transgenic plants.
© 2016 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures





References
-
- Andrés, F. and Coupland, G. (2012) The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. - PubMed
-
- Bäurle, I. and Dean, C. (2006) The timing of developmental transitions in plants. Cell, 125, 655–664. - PubMed
-
- Böhlenius, H. , Huang, T. , Charbonnel‐Campaa, L. , Brunner, A.M. , Jansson, S. , Strauss, S.H. and Nilsson, O. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science, 312, 1040–1043. - PubMed
-
- Chen, S. , Hawighorst, P. , Sun, J. and Polle, A. (2014) Salt tolerance in Populus: significance of stress signaling networks, mycorrhization, and soil amendments for cellular and whole‐plant nutrition. Environ. Exp. Bot. 107, 113–124.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

