Epstein-Barr viral microRNAs target caspase 3
- PMID: 27565721
- PMCID: PMC5002152
- DOI: 10.1186/s12985-016-0602-7
Epstein-Barr viral microRNAs target caspase 3
Abstract
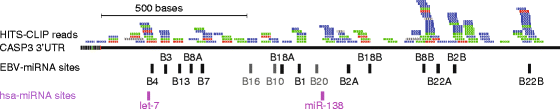
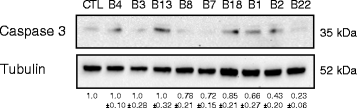
The Epstein-Barr virus (EBV) is a ubiquitous herpesvirus that transforms B cells and causes several malignancies including Burkitt's lymphoma. EBV differentially expresses at least 49 mature microRNAs (miRNAs) during latency in various infected epithelial and B cells. Recent high-throughput studies and functional assays have begun to reveal the function of the EBV miRNAs suggesting roles in latency, cell cycle control, and apoptosis. In particular, the central executioner of apoptosis, Caspase 3 (CASP3), was proposed as a target of select EBV miRNAs. However, whether CASP3 is truly a target of EBV miRNAs, and if so, which specific miRNAs target CASP3 is still under debate. Based on previously published high-throughput biochemical data and a bioinformatic analysis of the entire CASP3 3'-UTR, we identified 12 EBV miRNAs that have one or more seed binding sites in the CASP3 3'-UTR. We individually tested all 12 miRNAs for repression of CASP3 in luciferase reporter assays, and nine showed statistically significant (P < 0.001) repression of a full-length CASP3 reporter. Further, three EBV miRNAs, including BART22, exhibited repression of endogenous CASP3 protein. These data confirm that CASP3 is a direct target of specific EBV BART miRNAs.
Keywords: Burkitt’s lymphoma; Caspase 3; Epstein-Barr virus; microRNA.
Figures

References
-
- Nourse JP, Crooks P, Keane C, Nguyen-Van D, Mujaj S, Ross N, Jones K, Vari F, Han E, Trappe R, Fink S, Gandhi MK. Expression profiling of Epstein-Barr virus-encoded microRNAs from paraffin-embedded formalin-fixed primary Epstein-Barr virus-positive B-cell lymphoma samples. J Virol Methods. 2012;184:46–54. doi: 10.1016/j.jviromet.2012.05.005. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

