Diversification of both KIR and NKG2 natural killer cell receptor genes in macaques - implications for highly complex MHC-dependent regulation of natural killer cells
- PMID: 27565739
- PMCID: PMC5215213
- DOI: 10.1111/imm.12666
Diversification of both KIR and NKG2 natural killer cell receptor genes in macaques - implications for highly complex MHC-dependent regulation of natural killer cells
Abstract
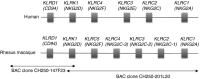
The killer immunoglobulin-like receptors (KIR) as well as their MHC class I ligands display enormous genetic diversity and polymorphism in macaque species. Signals resulting from interaction between KIR or CD94/NKG2 receptors and their cognate MHC class I proteins essentially regulate the activity of natural killer (NK) cells. Macaque and human KIR share many features, such as clonal expression patterns, gene copy number variations, specificity for particular MHC class I allotypes, or epistasis between KIR and MHC class I genes that influence susceptibility and resistance to immunodeficiency virus infection. In this review article we also annotated publicly available rhesus macaque BAC clone sequences and provide the first description of the CD94-NKG2 genomic region. Besides the presence of genes that are orthologous to human NKG2A and NKG2F, this region contains three NKG2C paralogues. Hence, the genome of rhesus macaques contains moderately expanded and diversified NKG2 genes in addition to highly diversified KIR genes. The presence of two diversified NK cell receptor families in one species has not been described before and is expected to require a complex MHC-dependent regulation of NK cells.
Keywords: MHC; genomics; killer immunoglobulin-like receptors; natural killer cells.
© 2016 John Wiley & Sons Ltd.
Figures

Similar articles
-
The KIR and CD94/NKG2 families of molecules in the rhesus monkey.Immunol Rev. 2001 Oct;183:25-40. doi: 10.1034/j.1600-065x.2001.1830103.x. Immunol Rev. 2001. PMID: 11782245 Review.
-
Two to Tango: Co-evolution of Hominid Natural Killer Cell Receptors and MHC.Front Immunol. 2019 Feb 19;10:177. doi: 10.3389/fimmu.2019.00177. eCollection 2019. Front Immunol. 2019. PMID: 30837985 Free PMC article. Review.
-
Structure and function of major histocompatibility complex (MHC) class I specific receptors expressed on human natural killer (NK) cells.Mol Immunol. 2002 Feb;38(9):637-60. doi: 10.1016/s0161-5890(01)00107-9. Mol Immunol. 2002. PMID: 11858820 Review.
-
HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C.Nature. 1998 Feb 19;391(6669):795-9. doi: 10.1038/35869. Nature. 1998. PMID: 9486650
-
NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C.J Immunol. 2002 Jul 1;169(1):220-9. doi: 10.4049/jimmunol.169.1.220. J Immunol. 2002. PMID: 12077248
Cited by
-
Phenotypic and Functional Characteristics of a Novel Influenza Virus Hemagglutinin-Specific Memory NK Cell.J Virol. 2021 May 24;95(12):e00165-21. doi: 10.1128/JVI.00165-21. Print 2021 May 24. J Virol. 2021. PMID: 33827945 Free PMC article.
-
MHC Class I Ligands of Rhesus Macaque Killer Cell Ig-like Receptors.J Immunol. 2023 Jun 1;210(11):1815-1826. doi: 10.4049/jimmunol.2200954. J Immunol. 2023. PMID: 37036309 Free PMC article.
-
Killer-cell immunoglobulin-like receptors on the cusp of modern immunogenetics.Immunology. 2017 Dec;152(4):556-561. doi: 10.1111/imm.12802. Epub 2017 Sep 12. Immunology. 2017. PMID: 28755388 Free PMC article. Review.
-
Detailed phenotypic and functional characterization of CMV-associated adaptive NK cells in rhesus macaques.Front Immunol. 2022 Nov 25;13:1028788. doi: 10.3389/fimmu.2022.1028788. eCollection 2022. Front Immunol. 2022. PMID: 36518759 Free PMC article.
-
Short tandem repeats, segmental duplications, gene deletion, and genomic instability in a rapidly diversified immune gene family.BMC Genomics. 2016 Nov 9;17(1):900. doi: 10.1186/s12864-016-3241-x. BMC Genomics. 2016. PMID: 27829352 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

