Binding-affinity predictions of HSP90 in the D3R Grand Challenge 2015 with docking, MM/GBSA, QM/MM, and free-energy simulations
- PMID: 27565797
- PMCID: PMC5078160
- DOI: 10.1007/s10822-016-9942-z
Binding-affinity predictions of HSP90 in the D3R Grand Challenge 2015 with docking, MM/GBSA, QM/MM, and free-energy simulations
Abstract
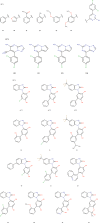
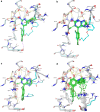
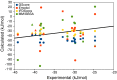
We have estimated the binding affinity of three sets of ligands of the heat-shock protein 90 in the D3R grand challenge blind test competition. We have employed four different methods, based on five different crystal structures: first, we docked the ligands to the proteins with induced-fit docking with the Glide software and calculated binding affinities with three energy functions. Second, the docked structures were minimised in a continuum solvent and binding affinities were calculated with the MM/GBSA method (molecular mechanics combined with generalised Born and solvent-accessible surface area solvation). Third, the docked structures were re-optimised by combined quantum mechanics and molecular mechanics (QM/MM) calculations. Then, interaction energies were calculated with quantum mechanical calculations employing 970-1160 atoms in a continuum solvent, combined with energy corrections for dispersion, zero-point energy and entropy, ligand distortion, ligand solvation, and an increase of the basis set to quadruple-zeta quality. Fourth, relative binding affinities were estimated by free-energy simulations, using the multi-state Bennett acceptance-ratio approach. Unfortunately, the results were varying and rather poor, with only one calculation giving a correlation to the experimental affinities larger than 0.7, and with no consistent difference in the quality of the predictions from the various methods. For one set of ligands, the results could be strongly improved (after experimental data were revealed) if it was recognised that one of the ligands displaced one or two water molecules. For the other two sets, the problem is probably that the ligands bind in different modes than in the crystal structures employed or that the conformation of the ligand-binding site or the whole protein changes.
Keywords: Bennett acceptance ratio; Big-QM; Blind-test competition; Continuum solvation; D3R grand challenge; Free-energy perturbation; Induced-fit docking; Ligand-binding affinity; MM/GBSA; QM/MM.
Figures
Similar articles
-
Large scale free energy calculations for blind predictions of protein-ligand binding: the D3R Grand Challenge 2015.J Comput Aided Mol Des. 2016 Sep;30(9):743-751. doi: 10.1007/s10822-016-9952-x. Epub 2016 Aug 25. J Comput Aided Mol Des. 2016. PMID: 27562018 Free PMC article.
-
Blinded predictions of binding modes and energies of HSP90-α ligands for the 2015 D3R grand challenge.Bioorg Med Chem. 2016 Oct 15;24(20):4890-4899. doi: 10.1016/j.bmc.2016.07.044. Epub 2016 Jul 21. Bioorg Med Chem. 2016. PMID: 27485604
-
Calculate protein-ligand binding affinities with the extended linear interaction energy method: application on the Cathepsin S set in the D3R Grand Challenge 3.J Comput Aided Mol Des. 2019 Jan;33(1):105-117. doi: 10.1007/s10822-018-0162-6. Epub 2018 Sep 14. J Comput Aided Mol Des. 2019. PMID: 30218199 Free PMC article.
-
The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities.Expert Opin Drug Discov. 2015 May;10(5):449-61. doi: 10.1517/17460441.2015.1032936. Epub 2015 Apr 2. Expert Opin Drug Discov. 2015. PMID: 25835573 Free PMC article. Review.
-
Ligand-Binding Affinity Estimates Supported by Quantum-Mechanical Methods.Chem Rev. 2016 May 11;116(9):5520-66. doi: 10.1021/acs.chemrev.5b00630. Epub 2016 Apr 14. Chem Rev. 2016. PMID: 27077817 Review.
Cited by
-
Docking of small molecules to farnesoid X receptors using AutoDock Vina with the Convex-PL potential: lessons learned from D3R Grand Challenge 2.J Comput Aided Mol Des. 2018 Jan;32(1):151-162. doi: 10.1007/s10822-017-0062-1. Epub 2017 Sep 14. J Comput Aided Mol Des. 2018. PMID: 28913782
-
DockingApp RF: A State-of-the-Art Novel Scoring Function for Molecular Docking in a User-Friendly Interface to AutoDock Vina.Int J Mol Sci. 2020 Dec 15;21(24):9548. doi: 10.3390/ijms21249548. Int J Mol Sci. 2020. PMID: 33333976 Free PMC article.
-
Quantum Chemical Approaches in Structure-Based Virtual Screening and Lead Optimization.Front Chem. 2018 May 29;6:188. doi: 10.3389/fchem.2018.00188. eCollection 2018. Front Chem. 2018. PMID: 29896472 Free PMC article. Review.
-
Identification of Drug Combination Therapies for SARS-CoV-2: A Molecular Dynamics Simulations Approach.Drug Des Devel Ther. 2022 Sep 9;16:2995-3013. doi: 10.2147/DDDT.S366423. eCollection 2022. Drug Des Devel Ther. 2022. PMID: 36110398 Free PMC article.
-
Binding affinities of the farnesoid X receptor in the D3R Grand Challenge 2 estimated by free-energy perturbation and docking.J Comput Aided Mol Des. 2018 Jan;32(1):211-224. doi: 10.1007/s10822-017-0056-z. Epub 2017 Sep 6. J Comput Aided Mol Des. 2018. PMID: 28879536 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

