Emerging metrology for high-throughput nanomaterial genotoxicology
- PMID: 27565834
- PMCID: PMC5500194
- DOI: 10.1093/mutage/gew037
Emerging metrology for high-throughput nanomaterial genotoxicology
Abstract
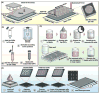
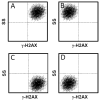
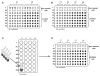
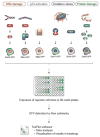
The rapid development of the engineered nanomaterial (ENM) manufacturing industry has accelerated the incorporation of ENMs into a wide variety of consumer products across the globe. Unintentionally or not, some of these ENMs may be introduced into the environment or come into contact with humans or other organisms resulting in unexpected biological effects. It is thus prudent to have rapid and robust analytical metrology in place that can be used to critically assess and/or predict the cytotoxicity, as well as the potential genotoxicity of these ENMs. Many of the traditional genotoxicity test methods [e.g. unscheduled DNA synthesis assay, bacterial reverse mutation (Ames) test, etc.,] for determining the DNA damaging potential of chemical and biological compounds are not suitable for the evaluation of ENMs, due to a variety of methodological issues ranging from potential assay interferences to problems centered on low sample throughput. Recently, a number of sensitive, high-throughput genotoxicity assays/platforms (CometChip assay, flow cytometry/micronucleus assay, flow cytometry/γ-H2AX assay, automated 'Fluorimetric Detection of Alkaline DNA Unwinding' (FADU) assay, ToxTracker reporter assay) have been developed, based on substantial modifications and enhancements of traditional genotoxicity assays. These new assays have been used for the rapid measurement of DNA damage (strand breaks), chromosomal damage (micronuclei) and for detecting upregulated DNA damage signalling pathways resulting from ENM exposures. In this critical review, we describe and discuss the fundamental measurement principles and measurement endpoints of these new assays, as well as the modes of operation, analytical metrics and potential interferences, as applicable to ENM exposures. An unbiased discussion of the major technical advantages and limitations of each assay for evaluating and predicting the genotoxic potential of ENMs is also provided.
Published by Oxford University Press on behalf of the UK Environmental Mutagen Society 2016.
Conflict of interest statement
Conflict of interest statement: G.H. is the CEO of Toxys, a Dutch biotech company that offers the ToxTracker assay as a commercial service to industry.
Figures

References
-
- Lux report. [accessed April 2016];Nanomaterials state of the market: Nanotechnology update: Corporations up their spending as revenues for nano-enabled products increase excerpt. https://portal.Luxresearchinc.Com/research/report_excerpt/16215.
-
- Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

