Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin
- PMID: 27566148
- PMCID: PMC5137432
- DOI: 10.1093/nar/gkw744
Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin
Abstract
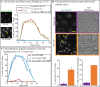
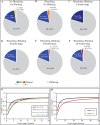
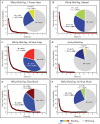
In vivo single molecule tracking has recently developed into a powerful technique for measuring and understanding the transient interactions of transcription factors (TF) with their chromatin response elements. However, this method still lacks a solid foundation for distinguishing between specific and non-specific interactions. To address this issue, we took advantage of the power of molecular genetics of yeast. Yeast TF Ace1p has only five specific sites in the genome and thus serves as a benchmark to distinguish specific from non-specific binding. Here, we show that the estimated residence time of the short-residence molecules is essentially the same for Hht1p, Ace1p and Hsf1p, equaling 0.12-0.32 s. These three DNA-binding proteins are very different in their structure, function and intracellular concentration. This suggests that (i) short-residence molecules are bound to DNA non-specifically, and (ii) that non-specific binding shares common characteristics between vastly different DNA-bound proteins and thus may have a common underlying mechanism. We develop new and robust procedure for evaluation of adverse effects of labeling, and new quantitative analysis procedures that significantly improve residence time measurements by accounting for fluorophore blinking. Our results provide a framework for the reliable performance and analysis of single molecule TF experiments in yeast.
Published by Oxford University Press on behalf of Nucleic Acids Research 2016. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures


References
-
- McNally J.G., Muller W.G., Walker D., Wolford R., Hager G.L. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. - PubMed
-
- Karpova T.S., Kim M.J., Spriet C., Nalley K., Stasevich T.J., Kherrouche Z., Heliot L., McNally J.G. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

