Using NMR and molecular dynamics to link structure and dynamics effects of the universal base 8-aza, 7-deaza, N8 linked adenosine analog
- PMID: 27566150
- PMCID: PMC5062995
- DOI: 10.1093/nar/gkw736
Using NMR and molecular dynamics to link structure and dynamics effects of the universal base 8-aza, 7-deaza, N8 linked adenosine analog
Abstract
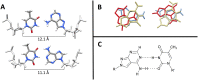
A truly universal nucleobase enables a host of novel applications such as simplified templates for PCR primers, randomized sequencing and DNA based devices. A universal base must pair indiscriminately to each of the canonical bases with little or preferably no destabilization of the overall duplex. In reality, many candidates either destabilize the duplex or do not base pair indiscriminatingly. The novel base 8-aza-7-deazaadenine (pyrazolo[3,4-d]pyrimidin- 4-amine) N8-(2'deoxyribonucleoside), a deoxyadenosine analog (UB), pairs with each of the natural DNA bases with little sequence preference. We have utilized NMR complemented with molecular dynamic calculations to characterize the structure and dynamics of a UB incorporated into a DNA duplex. The UB participates in base stacking with little to no perturbation of the local structure yet forms an unusual base pair that samples multiple conformations. These local dynamics result in the complete disappearance of a single UB proton resonance under native conditions. Accommodation of the UB is additionally stabilized via heightened backbone conformational sampling. NMR combined with various computational techniques has allowed for a comprehensive characterization of both structural and dynamic effects of the UB in a DNA duplex and underlines that the UB as a strong candidate for universal base applications.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
The N(8)-(2'-deoxyribofuranoside) of 8-aza-7-deazaadenine: a universal nucleoside forming specific hydrogen bonds with the four canonical DNA constituents.Nucleic Acids Res. 2000 Sep 1;28(17):3224-32. doi: 10.1093/nar/28.17.3224. Nucleic Acids Res. 2000. PMID: 10954589 Free PMC article.
-
Influence of PNA containing 8-aza-7-deazaadenine on structure stability and binding affinity of PNA·DNA duplex: insights from thermodynamics, counter ion, hydration and molecular dynamics analysis.Mol Biosyst. 2013 Jul;9(7):1958-71. doi: 10.1039/c3mb25561a. Epub 2013 May 1. Mol Biosyst. 2013. PMID: 23636232
-
Pyrazolo[3,4-d][1,2,3]triazine DNA: synthesis and base pairing of 7-deaza-2,8-diaza-2'-deoxyadenosine.J Org Chem. 2004 Jul 9;69(14):4695-700. doi: 10.1021/jo040150i. J Org Chem. 2004. PMID: 15230592
-
Azide-alkyne "click" conjugation of 8-aza-7-deazaadenine-DNA: synthesis, duplex stability, and fluorogenic dye labeling.Bioconjug Chem. 2010 Sep 15;21(9):1629-41. doi: 10.1021/bc100090y. Bioconjug Chem. 2010. PMID: 20681566
-
Chromophore/DNA interactions: femto- to nanosecond spectroscopy, NMR structure, and electron transfer theory.J Phys Chem B. 2008 Jan 24;112(3):973-89. doi: 10.1021/jp076405o. Epub 2007 Dec 29. J Phys Chem B. 2008. PMID: 18163608
References
-
- Hirao I., Kimoto M., Yamashige R. Natural versus artificial creation of base pairs in DNA: Origin of nucleobases from the perspectives of unnatural base pair studies. Acc. Chem. Res. 2012;45:2055–2065. - PubMed
-
- Kimoto M., Kawai R., Mitsui T., Yokoyama S., Hirao I. Efficient PCR amplification by an unnatural base pair system. Nucleic Acids Symp. Ser. (Oxf) 2008:469–470. - PubMed
-
- Chiaramonte M., Moore C.L., Kincaid K., Kuchta R.D. Facile polymerization of dNTPs bearing unnatural base analogues by DNA polymerase α and Klenow fragment (DNA polymerase I) Biochemistry. 2003;42:10472–10481. - PubMed
-
- Loakes D., Van Aerschotl A., Brown D.M., Hill F. Enzymatic recognition of acyclic universal base analogues in oligonucleotides. Nucleosides Nucleotides Nucleic Acids. 1996;15:1891–1904.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

