Clinical Actionability of Comprehensive Genomic Profiling for Management of Rare or Refractory Cancers
- PMID: 27566247
- PMCID: PMC5189630
- DOI: 10.1634/theoncologist.2016-0049
Clinical Actionability of Comprehensive Genomic Profiling for Management of Rare or Refractory Cancers
Abstract
Background: The frequency with which targeted tumor sequencing results will lead to implemented change in care is unclear. Prospective assessment of the feasibility and limitations of using genomic sequencing is critically important.
Methods: A prospective clinical study was conducted on 100 patients with diverse-histology, rare, or poor-prognosis cancers to evaluate the clinical actionability of a Clinical Laboratory Improvement Amendments (CLIA)-certified, comprehensive genomic profiling assay (FoundationOne), using formalin-fixed, paraffin-embedded tumors. The primary objectives were to assess utility, feasibility, and limitations of genomic sequencing for genomically guided therapy or other clinical purpose in the setting of a multidisciplinary molecular tumor board.
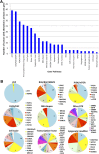
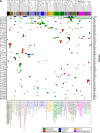
Results: Of the tumors from the 92 patients with sufficient tissue, 88 (96%) had at least one genomic alteration (average 3.6, range 0-10). Commonly altered pathways included p53 (46%), RAS/RAF/MAPK (rat sarcoma; rapidly accelerated fibrosarcoma; mitogen-activated protein kinase) (45%), receptor tyrosine kinases/ligand (44%), PI3K/AKT/mTOR (phosphatidylinositol-4,5-bisphosphate 3-kinase; protein kinase B; mammalian target of rapamycin) (35%), transcription factors/regulators (31%), and cell cycle regulators (30%). Many low frequency but potentially actionable alterations were identified in diverse histologies. Use of comprehensive profiling led to implementable clinical action in 35% of tumors with genomic alterations, including genomically guided therapy, diagnostic modification, and trigger for germline genetic testing.
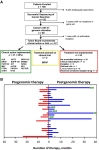
Conclusion: Use of targeted next-generation sequencing in the setting of an institutional molecular tumor board led to implementable clinical action in more than one third of patients with rare and poor-prognosis cancers. Major barriers to implementation of genomically guided therapy were clinical status of the patient and drug access. Early and serial sequencing in the clinical course and expanded access to genomically guided early-phase clinical trials and targeted agents may increase actionability.
Implications for practice: Identification of key factors that facilitate use of genomic tumor testing results and implementation of genomically guided therapy may lead to enhanced benefit for patients with rare or difficult to treat cancers. Clinical use of a targeted next-generation sequencing assay in the setting of an institutional molecular tumor board led to implementable clinical action in over one third of patients with rare and poor prognosis cancers. The major barriers to implementation of genomically guided therapy were clinical status of the patient and drug access both on trial and off label. Approaches to increase actionability include early and serial sequencing in the clinical course and expanded access to genomically guided early phase clinical trials and targeted agents.
Keywords: Cancer; Molecular sequencing; Molecular targeted therapy; Mutation; Tumor genomics.
©AlphaMed Press.
Conflict of interest statement
Disclosures of potential conflicts of interest may be found at the end of this article.
Figures


References
-
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. - PubMed
-
- Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

