Emerging facets of prokaryotic glycosylation
- PMID: 27566466
- PMCID: PMC5266552
- DOI: 10.1093/femsre/fuw036
Emerging facets of prokaryotic glycosylation
Abstract
Glycosylation of proteins is one of the most prevalent post-translational modifications occurring in nature, with a wide repertoire of biological implications. Pathways for the main types of this modification, the N- and O-glycosylation, can be found in all three domains of life-the Eukarya, Bacteria and Archaea-thereby following common principles, which are valid also for lipopolysaccharides, lipooligosaccharides and glycopolymers. Thus, studies on any glycoconjugate can unravel novel facets of the still incompletely understood fundamentals of protein N- and O-glycosylation. While it is estimated that more than two-thirds of all eukaryotic proteins would be glycosylated, no such estimate is available for prokaryotic glycoproteins, whose understanding is lagging behind, mainly due to the enormous variability of their glycan structures and variations in the underlying glycosylation processes. Combining glycan structural information with bioinformatic, genetic, biochemical and enzymatic data has opened up an avenue for in-depth analyses of glycosylation processes as a basis for glycoengineering endeavours. Here, the common themes of glycosylation are conceptualised for the major classes of prokaryotic (i.e. bacterial and archaeal) glycoconjugates, with a special focus on glycosylated cell-surface proteins. We describe the current knowledge of biosynthesis and importance of these glycoconjugates in selected pathogenic and beneficial microbes.
Keywords: glycan biosynthesis; glycoengineering; glycoproteins; prokaryotes; secondary cell-wall polymers; surface (S-) layer.
© FEMS 2016. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
. None declared.
Figures
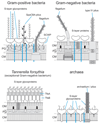
 , peptidoglycan strand; OM, outer membrane;
, peptidoglycan strand; OM, outer membrane;  , outer membrane lipid A;
, outer membrane lipid A;  , bacterial and archaeal membrane phospholipid;
, bacterial and archaeal membrane phospholipid;  , archaeal membrane tetraetherlipid;
, archaeal membrane tetraetherlipid;  ,
,  ,
,  , different S-layer glycans; SCWP, secondary cell wall polymer. In different archaeal species, S-layer (glyco)protein anchoring to the cell envelope has been suggested either by a protein transmembrane anchor
, different S-layer glycans; SCWP, secondary cell wall polymer. In different archaeal species, S-layer (glyco)protein anchoring to the cell envelope has been suggested either by a protein transmembrane anchor  (Lechner and Wieland 1989) or in an archaeosortase-dependent process by a lipid anchor
(Lechner and Wieland 1989) or in an archaeosortase-dependent process by a lipid anchor  (Abdul Halim et al. 2016). (Extended and modified from Messner, Schäffer and Kosma 2013. With permission from Elsevier).
(Abdul Halim et al. 2016). (Extended and modified from Messner, Schäffer and Kosma 2013. With permission from Elsevier).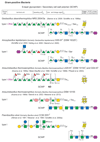
 , hexuronic acid;
, hexuronic acid; , glucose;
, glucose; , N-acetylglucosamine;
, N-acetylglucosamine; , glucuronic acid;
, glucuronic acid; , 3-N-acetylquinovosamine;
, 3-N-acetylquinovosamine; , 2,3-di-N-acetylglucuronic acid;
, 2,3-di-N-acetylglucuronic acid; , 6-sulfoquinovose;
, 6-sulfoquinovose; , 2-amino-6-sulfo-2,6-dideoxy-quinovose;
, 2-amino-6-sulfo-2,6-dideoxy-quinovose; , galactose;
, galactose; , galactofuranose;
, galactofuranose; , N-acetylgalactosamine;
, N-acetylgalactosamine; , galacturonic acid;
, galacturonic acid; , 3-O-methylgalacturonic acid;
, 3-O-methylgalacturonic acid; , mannose;
, mannose; , N-acetylmannosamine;
, N-acetylmannosamine; , mannosaminuronic acid;
, mannosaminuronic acid; , 6-N-amido-Thr mannosaminuronic acid;
, 6-N-amido-Thr mannosaminuronic acid; ,
,  ,
,  ,
,  ,
,  , 3-N-acetylfucosamine;
, 3-N-acetylfucosamine; , iduronic acid;
, iduronic acid; , ribose;
, ribose; ,
,  , xylose;
, xylose; , digitoxose;
, digitoxose; , 5-acetamidino-7-glycerolyl pseudaminic acid;
, 5-acetamidino-7-glycerolyl pseudaminic acid; , N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
, N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
 , hexuronic acid;
, hexuronic acid; , glucose;
, glucose; , N-acetylglucosamine;
, N-acetylglucosamine; , glucuronic acid;
, glucuronic acid; , 3-N-acetylquinovosamine;
, 3-N-acetylquinovosamine; , 2,3-di-N-acetylglucuronic acid;
, 2,3-di-N-acetylglucuronic acid; , 6-sulfoquinovose;
, 6-sulfoquinovose; , 2-amino-6-sulfo-2,6-dideoxy-quinovose;
, 2-amino-6-sulfo-2,6-dideoxy-quinovose; , galactose;
, galactose; , galactofuranose;
, galactofuranose; , N-acetylgalactosamine;
, N-acetylgalactosamine; , galacturonic acid;
, galacturonic acid; , 3-O-methylgalacturonic acid;
, 3-O-methylgalacturonic acid; , mannose;
, mannose; , N-acetylmannosamine;
, N-acetylmannosamine; , mannosaminuronic acid;
, mannosaminuronic acid; , 6-N-amido-Thr mannosaminuronic acid;
, 6-N-amido-Thr mannosaminuronic acid; ,
,  ,
,  ,
,  ,
,  , 3-N-acetylfucosamine;
, 3-N-acetylfucosamine; , iduronic acid;
, iduronic acid; , ribose;
, ribose; ,
,  , xylose;
, xylose; , digitoxose;
, digitoxose; , 5-acetamidino-7-glycerolyl pseudaminic acid;
, 5-acetamidino-7-glycerolyl pseudaminic acid; , N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
, N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
 , hexuronic acid;
, hexuronic acid; , glucose;
, glucose; , N-acetylglucosamine;
, N-acetylglucosamine; , glucuronic acid;
, glucuronic acid; , 3-N-acetylquinovosamine;
, 3-N-acetylquinovosamine; , 2,3-di-N-acetylglucuronic acid;
, 2,3-di-N-acetylglucuronic acid; , 6-sulfoquinovose;
, 6-sulfoquinovose; , 2-amino-6-sulfo-2,6-dideoxy-quinovose;
, 2-amino-6-sulfo-2,6-dideoxy-quinovose; , galactose;
, galactose; , galactofuranose;
, galactofuranose; , N-acetylgalactosamine;
, N-acetylgalactosamine; , galacturonic acid;
, galacturonic acid; , 3-O-methylgalacturonic acid;
, 3-O-methylgalacturonic acid; , mannose;
, mannose; , N-acetylmannosamine;
, N-acetylmannosamine; , mannosaminuronic acid;
, mannosaminuronic acid; , 6-N-amido-Thr mannosaminuronic acid;
, 6-N-amido-Thr mannosaminuronic acid; ,
,  ,
,  ,
,  ,
,  , 3-N-acetylfucosamine;
, 3-N-acetylfucosamine; , iduronic acid;
, iduronic acid; , ribose;
, ribose; ,
,  , xylose;
, xylose; , digitoxose;
, digitoxose; , 5-acetamidino-7-glycerolyl pseudaminic acid;
, 5-acetamidino-7-glycerolyl pseudaminic acid; , N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
, N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
 , hexuronic acid;
, hexuronic acid; , glucose;
, glucose; , N-acetylglucosamine;
, N-acetylglucosamine; , glucuronic acid;
, glucuronic acid; , 3-N-acetylquinovosamine;
, 3-N-acetylquinovosamine; , 2,3-di-N-acetylglucuronic acid;
, 2,3-di-N-acetylglucuronic acid; , 6-sulfoquinovose;
, 6-sulfoquinovose; , 2-amino-6-sulfo-2,6-dideoxy-quinovose;
, 2-amino-6-sulfo-2,6-dideoxy-quinovose; , galactose;
, galactose; , galactofuranose;
, galactofuranose; , N-acetylgalactosamine;
, N-acetylgalactosamine; , galacturonic acid;
, galacturonic acid; , 3-O-methylgalacturonic acid;
, 3-O-methylgalacturonic acid; , mannose;
, mannose; , N-acetylmannosamine;
, N-acetylmannosamine; , mannosaminuronic acid;
, mannosaminuronic acid; , 6-N-amido-Thr mannosaminuronic acid;
, 6-N-amido-Thr mannosaminuronic acid; ,
,  ,
,  ,
,  ,
,  , 3-N-acetylfucosamine;
, 3-N-acetylfucosamine; , iduronic acid;
, iduronic acid; , ribose;
, ribose; ,
,  , xylose;
, xylose; , digitoxose;
, digitoxose; , 5-acetamidino-7-glycerolyl pseudaminic acid;
, 5-acetamidino-7-glycerolyl pseudaminic acid; , N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).
, N-acetylmuramic acid. Monosaccharide symbols follow the SNFG (Symbol Nomenclature for Glycans) (Appendix-1B 2015). (Mescher and Strominger 1976; Koval and Jarrell 1987; Lechner and Sumper 1987; Sumper et al. 1990; Altman et al. 1991, 1992, 1995, 1996; Baumeister and Lembcke 1992; Christian et al. 1993; Kärcher et al. 1993; Möschl et al. 1993; Bock et al. 1994; Messner et al. 1995; Kosma et al. 1995a,b; Meier-Stauffer et al. 1996; Schäffer et al. 1999a,b, 2000a,b, 2004; Eichler 2000, 2013; Steindl et al. 2002, 2005; Pfoestl et al. 2003; Kählig et al. 2005; Voisin et al. 2005; Steiner et al. 2008; Veith et al. 2009; Peyfoon et al. 2010; Posch et al. 2011; Messner, Schäffer and Kosma 2013; Palmieri et al. 2013; Kandiba and Eichler 2014; Parente et al. 2014; Anzengruber et al. 2014a; Appendix-1B 2015; Lu et al. 2015; Kandiba et al. 2016). (Extended and modified from Eichler 2013. With permission from Nature Publishing Group).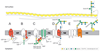
References
-
- Abdul Halim MF, Pfeiffer F, Zou J, et al. Haloferax volcanii archaeosortase is required for motility, mating, and C-terminal processing of the S-layer glycoprotein. Mol Microbiol. 2013;88:1164–75. - PubMed
-
- Abu-Qarn M, Yurist-Doutsch S, Giordano A, et al. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J Mol Biol. 2007;374:1224–36. - PubMed
-
- Adamo R. Glycan surface antigens from Bacillus anthracis as vaccine targets: current status and future perspectives. Expert Rev Vaccines. 2014;13:895–907. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

