Three-Dimensional Digital Template Atlas of the Macaque Brain
- PMID: 27566980
- PMCID: PMC6075609
- DOI: 10.1093/cercor/bhw248
Three-Dimensional Digital Template Atlas of the Macaque Brain
Abstract
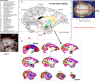
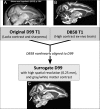
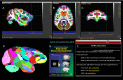
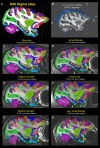
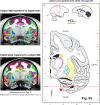
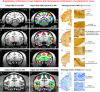
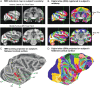
We present a new 3D template atlas of the anatomical subdivisions of the macaque brain, which is based on and aligned to the magnetic resonance imaging (MRI) data set and histological sections of the Saleem and Logothetis atlas. We describe the creation and validation of the atlas that, when registered with macaque structural or functional MRI scans, provides a straightforward means to estimate the boundaries between architectonic areas, either in a 3D volume with different planes of sections, or on an inflated brain surface (cortical flat map). As such, this new template atlas is intended for use as a reference standard for macaque brain research. Atlases and templates are available as both volumes and surfaces in standard NIFTI and GIFTI formats.
Keywords: 3D digital atlas; AFNI and SUMA; anatomical templates; architectonic areas; macaque monkey.
Published by Oxford University Press 2016.
Figures

References
-
- Bezgin G, Vakorin VA, van Opstal AJ, McIntosh AR, Bakker R. 2012. Hundreds of brain maps in one atlas: registering coordinate-independent primate neuro-anatomical data to a standard brain. Neuroimage. 62:67–76. - PubMed
-
- Brodmann K. 1909. Vergleichende Lokalisationslehre der Grosshirnrinde. Leipzig: Johann Ambrosius Barth.
-
- Chakravarty MM, Sadikot AF, Germann J, Bertrand G, Collins DL. 2008. Towards a validation of atlas warping techniques. Med Image Anal. 12:713–726. - PubMed
-
- Chakravarty MM, Frey S, Collins DL. 2009. Digital atlas of the rhesus monkey brain in stereotaxic coordinates In: Paxinos G, Huang X-F, Petrides M, Toga AW, editors. The rhesus monkey brain in stereotaxic coordinates. San Diego: Academic Press; p 403–407.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

