Lessons from Retinoblastoma: Implications for Cancer, Development, Evolution, and Regenerative Medicine
- PMID: 27567287
- PMCID: PMC5048577
- DOI: 10.1016/j.molmed.2016.07.010
Lessons from Retinoblastoma: Implications for Cancer, Development, Evolution, and Regenerative Medicine
Abstract
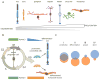
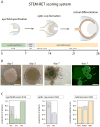
Retinoblastoma is a rare childhood cancer of the developing retina, and studies on this orphan disease have led to fundamental discoveries in cancer biology. Retinoblastoma has also emerged as a model for translational research for pediatric solid tumors, which is particularly important as personalized medicine expands in oncology. Research on retinoblastomas has been combined with the exploration of retinal development and retinal degeneration to advance a new model of cell type-specific disease susceptibility termed 'cellular pliancy'. The concept can even be extended to species-specific regeneration. This review discusses the remarkable path of retinoblastoma research and how it has shaped the most current efforts in basic, translational, and clinical research in oncology and beyond.
Keywords: cellular pliancy; orthotopic patient-derived xenografts; pediatric solid tumors; retinal regeneration; retinoblastoma.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Retinoblastoma: An update.Semin Diagn Pathol. 2016 May;33(3):133-40. doi: 10.1053/j.semdp.2015.10.007. Epub 2015 Oct 23. Semin Diagn Pathol. 2016. PMID: 26969537
-
Retinoblastoma: animal models.Ophthalmol Clin North Am. 2005 Mar;18(1):25-39, vii. doi: 10.1016/j.ohc.2004.08.006. Ophthalmol Clin North Am. 2005. PMID: 15763189 Review.
-
Retinoblastoma, the visible CNS tumor: A review.J Neurosci Res. 2019 Jan;97(1):29-44. doi: 10.1002/jnr.24213. Epub 2018 Jan 3. J Neurosci Res. 2019. PMID: 29314142 Free PMC article. Review.
-
pRB-Depleted Pluripotent Stem Cell Retinal Organoids Recapitulate Cell State Transitions of Retinoblastoma Development and Suggest an Important Role for pRB in Retinal Cell Differentiation.Stem Cells Transl Med. 2022 Apr 29;11(4):415-433. doi: 10.1093/stcltm/szac008. Stem Cells Transl Med. 2022. PMID: 35325233 Free PMC article.
-
Retinoblastoma.Pediatr Clin North Am. 2015 Feb;62(1):201-23. doi: 10.1016/j.pcl.2014.09.014. Pediatr Clin North Am. 2015. PMID: 25435120 Review.
Cited by
-
A short-term chick embryo in vivo xenograft model to study retinoblastoma cancer stem cells.Indian J Ophthalmol. 2022 May;70(5):1703-1711. doi: 10.4103/ijo.IJO_2348_21. Indian J Ophthalmol. 2022. PMID: 35502056 Free PMC article.
-
The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis.Neuron. 2017 May 3;94(3):550-568.e10. doi: 10.1016/j.neuron.2017.04.022. Neuron. 2017. PMID: 28472656 Free PMC article.
-
Characterization and Molecular Mechanism of Peptide-Conjugated Gold Nanoparticle Inhibiting p53-HDM2 Interaction in Retinoblastoma.Mol Ther Nucleic Acids. 2017 Dec 15;9:349-364. doi: 10.1016/j.omtn.2017.10.012. Epub 2017 Oct 20. Mol Ther Nucleic Acids. 2017. PMID: 29246314 Free PMC article.
-
Dissecting the Transcriptional and Chromatin Accessibility Heterogeneity of Proliferating Cone Precursors in Human Retinoblastoma Tumors by Single Cell Sequencing-Opening Pathways to New Therapeutic Strategies?Invest Ophthalmol Vis Sci. 2021 May 3;62(6):18. doi: 10.1167/iovs.62.6.18. Invest Ophthalmol Vis Sci. 2021. PMID: 34003213 Free PMC article.
-
Pentoxifylline Enhances the Apoptotic Effect of Carboplatin in Y79 Retinoblastoma Cells.In Vivo. 2019 Mar-Apr;33(2):401-412. doi: 10.21873/invivo.11487. In Vivo. 2019. PMID: 30804118 Free PMC article.
References
-
- Rodriguez-Galindo C, et al. Retinoblastoma. In: Orkin S, editor. Hematology and Oncology of Infancy and Childhood. Elsevier; 2015. pp. 1747–1778.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

