Microarray-based identification of genes associated with cancer progression and prognosis in hepatocellular carcinoma
- PMID: 27567667
- PMCID: PMC5002170
- DOI: 10.1186/s13046-016-0403-2
Microarray-based identification of genes associated with cancer progression and prognosis in hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths. The average survival and 5-year survival rates of HCC patients still remains poor. Thus, there is an urgent need to better understand the mechanisms of cancer progression in HCC and to identify useful biomarkers to predict prognosis.
Methods: Public data portals including Oncomine, The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) profiles were used to retrieve the HCC-related microarrays and to identify potential genes contributed to cancer progression. Bioinformatics analyses including pathway enrichment, protein/gene interaction and text mining were used to explain the potential roles of the identified genes in HCC. Quantitative real-time polymerase chain reaction analysis and Western blotting were used to measure the expression of the targets. The data were analysed by SPSS 20.0 software.
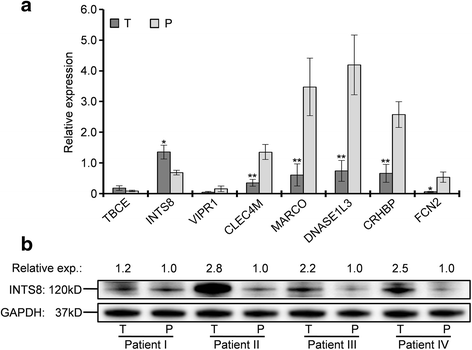
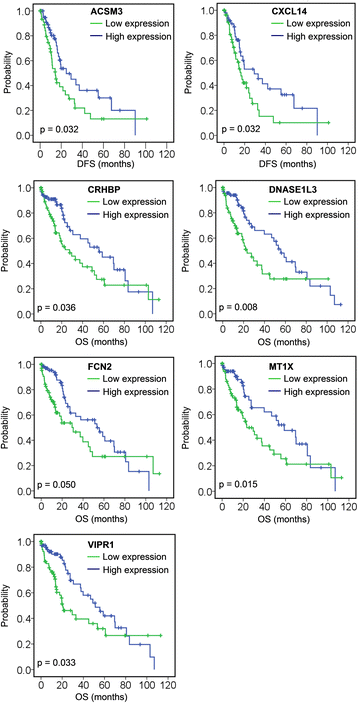
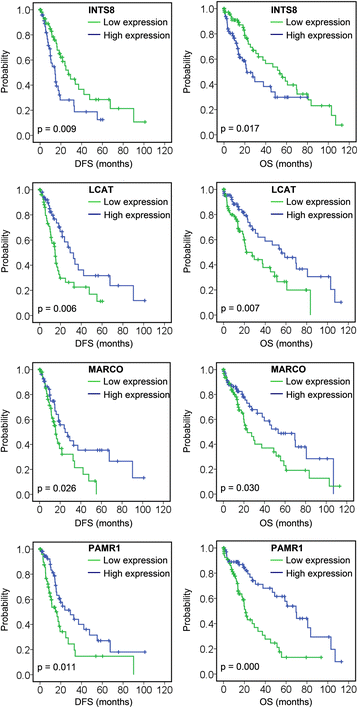
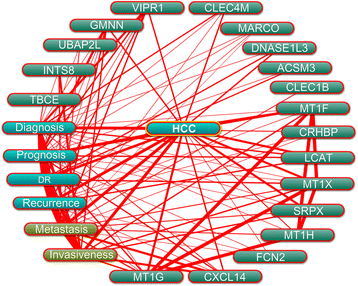
Results: We identified 80 genes that were significantly dysregulated in HCC according to four independent microarrays covering 386 cases of HCC and 327 normal liver tissues. Twenty genes were consistently and stably dysregulated in the four microarrays by at least 2-fold and detection of gene expression by RT-qPCR and western blotting showed consistent expression profiles in 11 HCC tissues compared with corresponding paracancerous tissues. Eleven of these 20 genes were associated with disease-free survival (DFS) or overall survival (OS) in a cohort of 157 HCC patients, and eight genes were associated with tumour pathologic PT, tumour stage or vital status. Potential roles of those 20 genes in regulation of HCC progression were predicted, primarily in association with metastasis. INTS8 was specifically correlated with most clinical characteristics including DFS, OS, stage, metastasis, invasiveness, diagnosis, and age.
Conclusion: The significantly dysregulated genes identified in this study were associated with cancer progression and prognosis in HCC, and might be potential therapeutic targets for HCC treatment or potential biomarkers for diagnosis and prognosis.
Keywords: Hepatocellular carcinoma; Microarray; Prognosis; Progression.
Figures



Similar articles
-
Analysis of microarrays of miR-34a and its identification of prospective target gene signature in hepatocellular carcinoma.BMC Cancer. 2018 Jan 3;18(1):12. doi: 10.1186/s12885-017-3941-x. BMC Cancer. 2018. PMID: 29298665 Free PMC article.
-
Expression of the Long Intergenic Non-Protein Coding RNA 665 (LINC00665) Gene and the Cell Cycle in Hepatocellular Carcinoma Using The Cancer Genome Atlas, the Gene Expression Omnibus, and Quantitative Real-Time Polymerase Chain Reaction.Med Sci Monit. 2018 May 5;24:2786-2808. doi: 10.12659/MSM.907389. Med Sci Monit. 2018. PMID: 29728556 Free PMC article.
-
SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma.Tumour Biol. 2016 Feb;37(2):2379-85. doi: 10.1007/s13277-015-4052-4. Epub 2015 Sep 16. Tumour Biol. 2016. PMID: 26373735
-
Loss of alanine-glyoxylate and serine-pyruvate aminotransferase expression accelerated the progression of hepatocellular carcinoma and predicted poor prognosis.J Transl Med. 2019 Nov 26;17(1):390. doi: 10.1186/s12967-019-02138-5. J Transl Med. 2019. PMID: 31771612 Free PMC article.
-
Long non-coding RNAs as biomarkers and therapeutic targets: Recent insights into hepatocellular carcinoma.Life Sci. 2017 Dec 15;191:273-282. doi: 10.1016/j.lfs.2017.10.007. Epub 2017 Oct 4. Life Sci. 2017. PMID: 28987633 Review.
Cited by
-
Classification of early and late stage liver hepatocellular carcinoma patients from their genomics and epigenomics profiles.PLoS One. 2019 Sep 6;14(9):e0221476. doi: 10.1371/journal.pone.0221476. eCollection 2019. PLoS One. 2019. PMID: 31490960 Free PMC article.
-
MicroRNA-31-5p regulates chemosensitivity by preventing the nuclear location of PARP1 in hepatocellular carcinoma.J Exp Clin Cancer Res. 2018 Nov 6;37(1):268. doi: 10.1186/s13046-018-0930-0. J Exp Clin Cancer Res. 2018. Retraction in: J Exp Clin Cancer Res. 2019 Apr 2;38(1):144. doi: 10.1186/s13046-019-1158-3. PMID: 30400960 Free PMC article. Retracted.
-
Microarray analyses reveal genes related to progression and prognosis of esophageal squamous cell carcinoma.Oncotarget. 2017 Aug 12;8(45):78838-78850. doi: 10.18632/oncotarget.20232. eCollection 2017 Oct 3. Oncotarget. 2017. PMID: 29108269 Free PMC article.
-
Predicting the Clinical Outcome of Lung Adenocarcinoma Using a Novel Gene Pair Signature Related to RNA-Binding Protein.Biomed Res Int. 2020 Oct 26;2020:8896511. doi: 10.1155/2020/8896511. eCollection 2020. Biomed Res Int. 2020. PMID: 33195699 Free PMC article.
-
SPP1 functions as an enhancer of cell growth in hepatocellular carcinoma targeted by miR-181c.Am J Transl Res. 2019 Nov 15;11(11):6924-6937. eCollection 2019. Am J Transl Res. 2019. PMID: 31814897 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

