Global Conservation of Protein Status between Cell Lines and Xenografts
- PMID: 27567954
- PMCID: PMC5006813
- DOI: 10.1016/j.tranon.2016.05.005
Global Conservation of Protein Status between Cell Lines and Xenografts
Abstract
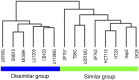
Common preclinical models for testing anticancer treatment include cultured human tumor cell lines in monolayer, and xenografts derived from these cell lines in immunodeficient mice. Our goal was to determine how similar the xenografts are compared with their original cell line and to determine whether it is possible to predict the stability of a xenograft model beforehand. We studied a selection of 89 protein markers of interest in 14 human cell cultures and respective subcutaneous xenografts using the reverse-phase protein array technology. We specifically focused on proteins and posttranslational modifications involved in DNA repair, PI3K pathway, apoptosis, tyrosine kinase signaling, stress, cell cycle, MAPK/ERK signaling, SAPK/JNK signaling, NFκB signaling, and adhesion/cytoskeleton. Using hierarchical clustering, most cell culture-xenograft pairs cluster together, suggesting a global conservation of protein signature. Particularly, Akt, NFkB, EGFR, and Vimentin showed very stable protein expression and phosphorylation levels highlighting that 4 of 10 pathways were highly correlated whatever the model. Other proteins were heterogeneously conserved depending on the cell line. Finally, cell line models with low Akt pathway activation and low levels of Vimentin gave rise to more reliable xenograft models. These results may be useful for the extrapolation of cell culture experiments to in vivo models in novel targeted drug discovery.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Stylli SS, Luwor RB, Ware TMB, Tan F, Kaye AH. Mouse models of glioma. J Clin Neurosci. 2015;22:619–626. - PubMed
-
- Marx V. Models: stretching the skills of cell lines and mice. Nat Methods. 2014;11:617–620. - PubMed
-
- Hickman JA, Graeser R, de Hoogt R, Vidic S, Brito C, Gutekunst M, van der Kuip H. IMI PREDECT Consortium. Three-dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol J. 2014;9:1115–1128. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

